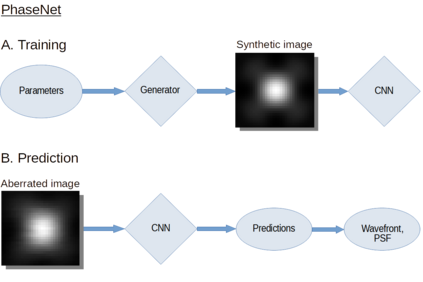
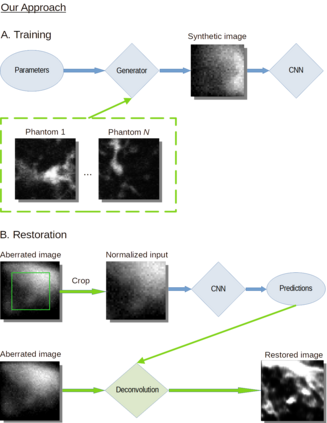
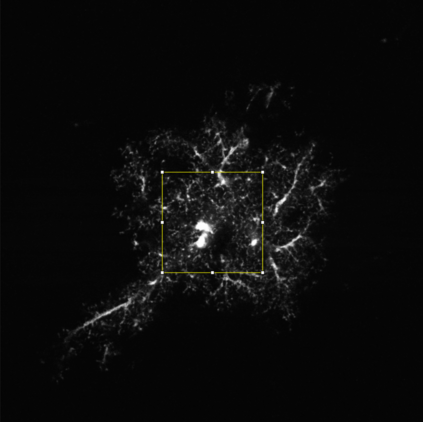
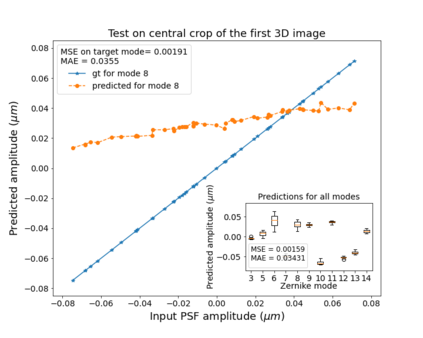
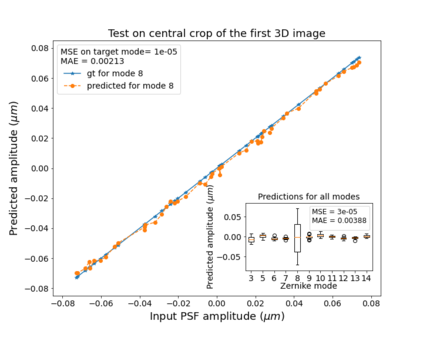
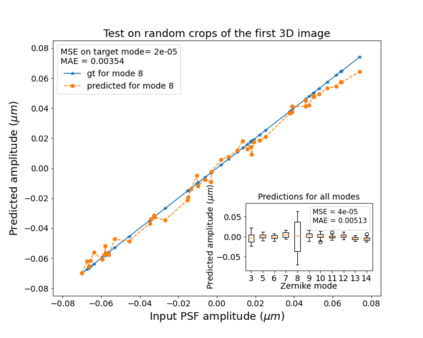
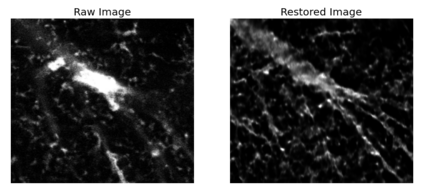
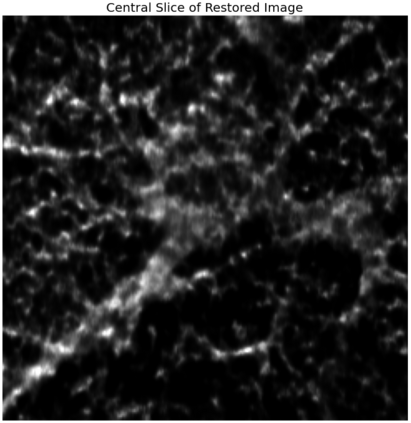
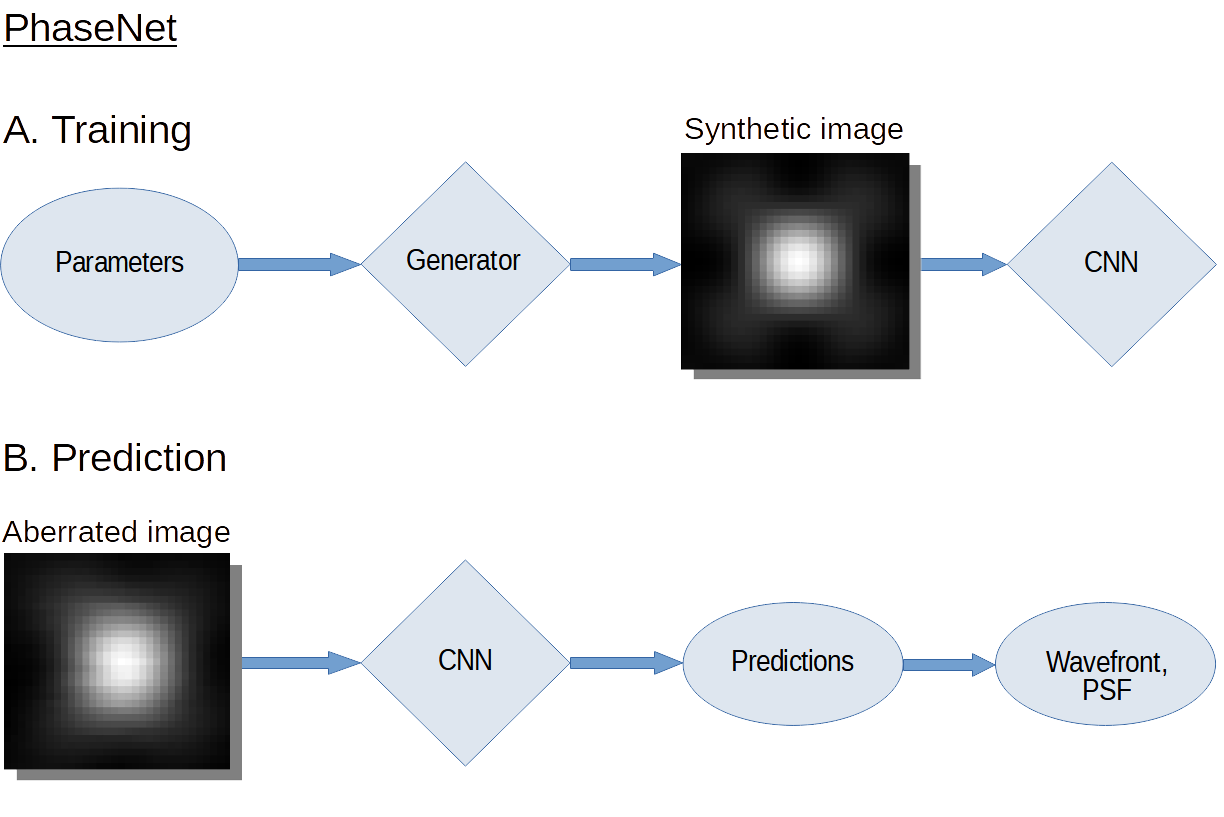
The quality of microscopy images often suffers from optical aberrations. These aberrations and their associated point spread functions have to be quantitatively estimated to restore aberrated images. The recent state-of-the-art method PhaseNet, based on a convolutional neural network, can quantify aberrations accurately but is limited to images of point light sources, e.g. fluorescent beads. In this research, we describe an extension of PhaseNet enabling its use on 3D images of biological samples. To this end, our method incorporates object-specific information into the simulated images used for training the network. Further, we add a Python-based restoration of images via Richardson-Lucy deconvolution. We demonstrate that the deconvolution with the predicted PSF can not only remove the simulated aberrations but also improve the quality of the real raw microscopic images with unknown residual PSF. We provide code for fast and convenient prediction and correction of aberrations.
翻译:显微镜像的质量往往受到光学偏差的影响。 这些偏差及其相关的点扩展功能必须量化估算才能恢复偏差图像。 最近的先进方法阶段网基于一个进化神经网络,可以准确量化偏差,但仅限于点光源(例如荧光珠)的图像。 在这个研究中, 我们描述SqualNet的扩展, 使其能够用于3D生物样本的图像。 为此, 我们的方法将特定对象的信息纳入用于培训网络的模拟图像中。 此外, 我们通过Richardson- Lucy de convolution, 添加了基于Python的图像恢复功能。 我们证明, 与预测的 PSF 的变异不仅可以去除模拟的偏差, 还可以提高用未知的残余 PSF 真实原始微谱图像的质量。 我们为快速和方便地预测和纠正异常提供了代码 。