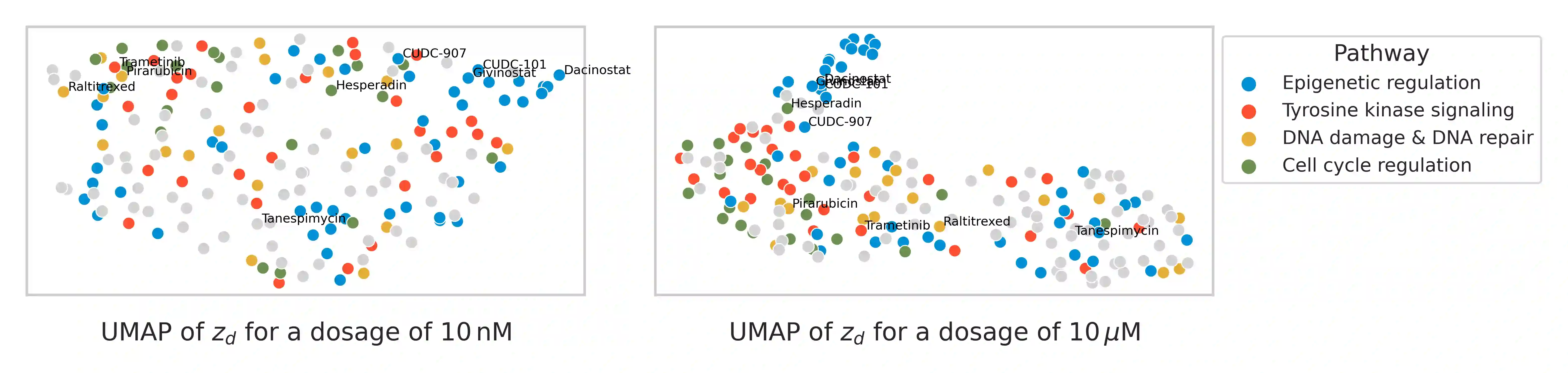
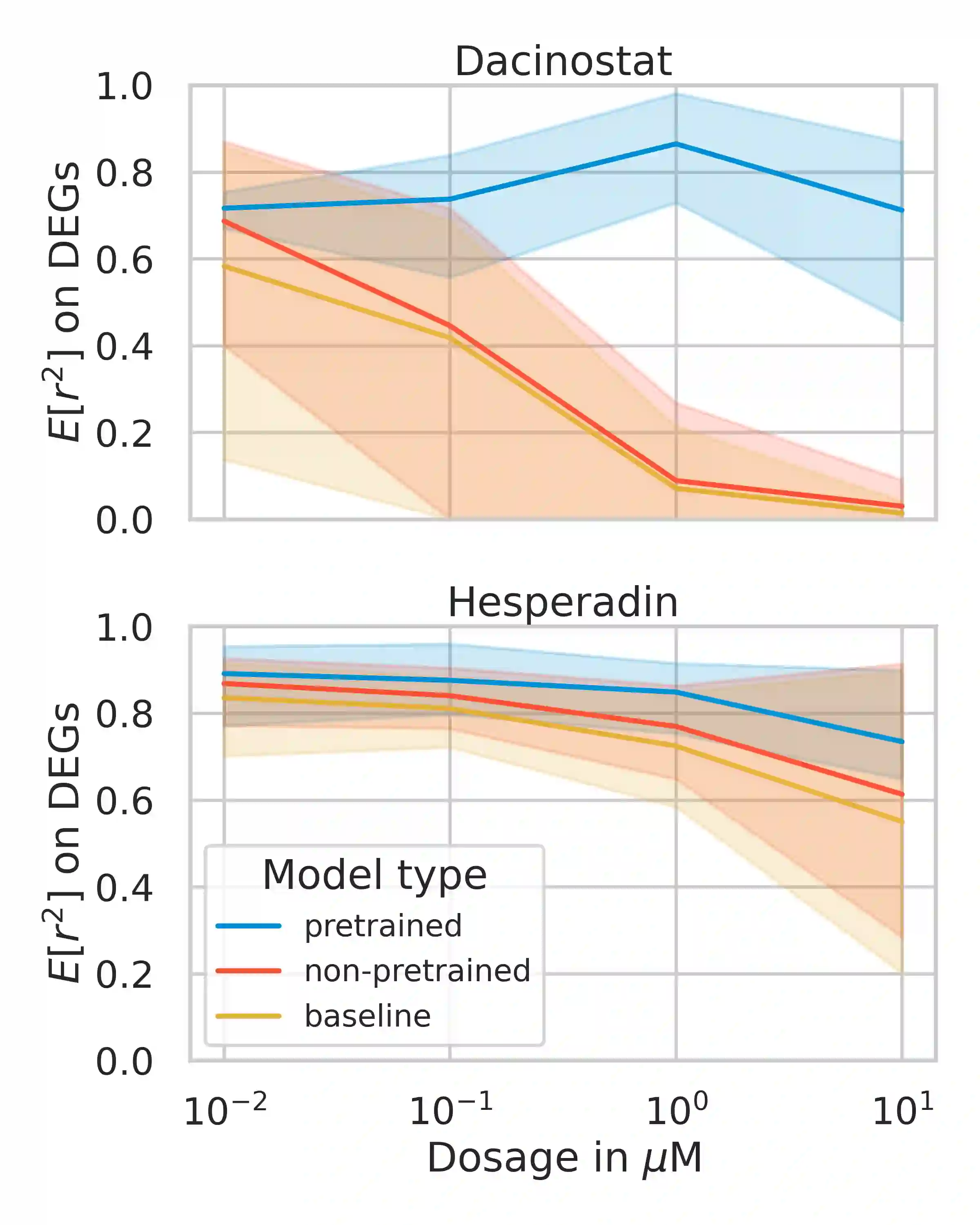
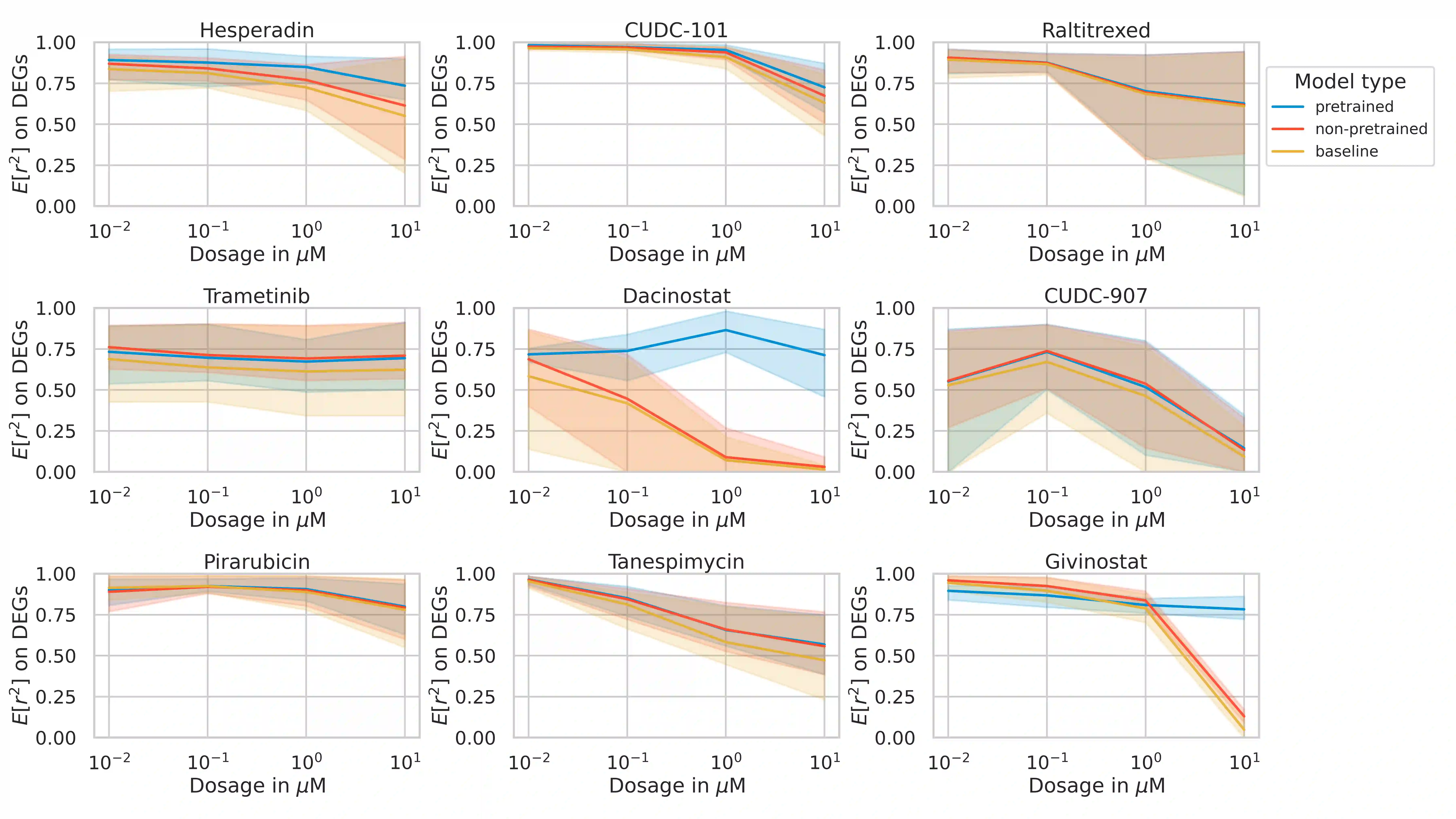
Single-cell transcriptomics enabled the study of cellular heterogeneity in response to perturbations at the resolution of individual cells. However, scaling high-throughput screens (HTSs) to measure cellular responses for many drugs remains a challenge due to technical limitations and, more importantly, the cost of such multiplexed experiments. Thus, transferring information from routinely performed bulk RNA-seq HTS is required to enrich single-cell data meaningfully. We introduce a new encoder-decoder architecture to study the perturbational effects of unseen drugs. We combine the model with a transfer learning scheme and demonstrate how training on existing bulk RNA-seq HTS datasets can improve generalisation performance. Better generalisation reduces the need for extensive and costly screens at single-cell resolution. We envision that our proposed method will facilitate more efficient experiment designs through its ability to generate in-silico hypotheses, ultimately accelerating targeted drug discovery.
翻译:单细胞转基因组学使得能够针对个别细胞的分解而研究细胞异质性,然而,由于技术限制,更重要的是,由于这种多轴实验的成本,衡量许多药物的细胞反应所需的高通量屏幕(HTS)规模仍然是一项挑战。因此,需要从例行进行的大型RNA-Seq HTS中传输信息,以有意义地丰富单细胞数据。我们引入了新的编码器脱coder结构,以研究未见药物的扰动效应。我们将该模型与转移学习计划结合起来,并展示如何对现有成批RNA-seq HTS数据集进行培训,以提高一般化绩效。更好的概括化减少了单细胞分辨率对广泛和昂贵的屏幕的需求。我们设想,我们拟议的方法将促进更有效的实验设计,因为它能够产生硅内假肢,最终加速定向药物发现。
相关内容
- Today (iOS and OS X): widgets for the Today view of Notification Center
- Share (iOS and OS X): post content to web services or share content with others
- Actions (iOS and OS X): app extensions to view or manipulate inside another app
- Photo Editing (iOS): edit a photo or video in Apple's Photos app with extensions from a third-party apps
- Finder Sync (OS X): remote file storage in the Finder with support for Finder content annotation
- Storage Provider (iOS): an interface between files inside an app and other apps on a user's device
- Custom Keyboard (iOS): system-wide alternative keyboards
Source: iOS 8 Extensions: Apple’s Plan for a Powerful App Ecosystem