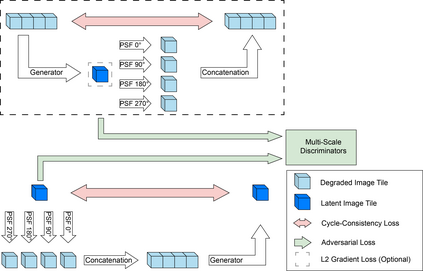
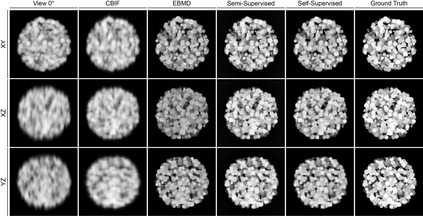
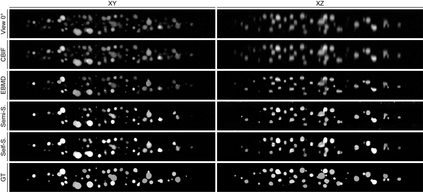
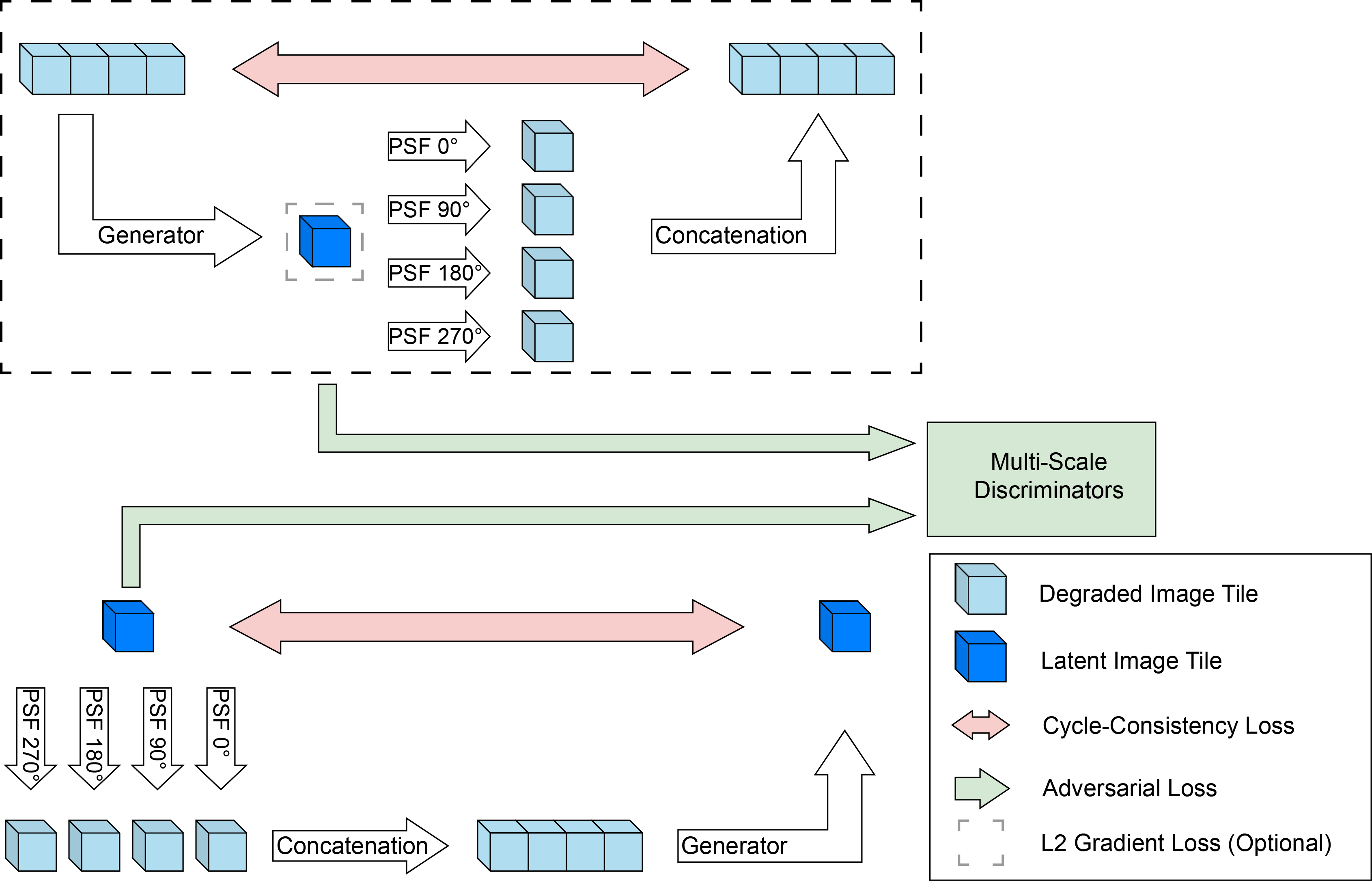
Recent developments in fluorescence microscopy allow capturing high-resolution 3D images over time for living model organisms. To be able to image even large specimens, techniques like multi-view light-sheet imaging record different orientations at each time point that can then be fused into a single high-quality volume. Based on measured point spread functions (PSF), deconvolution and content fusion are able to largely revert the inevitable degradation occurring during the imaging process. Classical multi-view deconvolution and fusion methods mainly use iterative procedures and content-based averaging. Lately, Convolutional Neural Networks (CNNs) have been deployed to approach 3D single-view deconvolution microscopy, but the multi-view case waits to be studied. We investigated the efficacy of CNN-based multi-view deconvolution and fusion with two synthetic data sets that mimic developing embryos and involve either two or four complementary 3D views. Compared with classical state-of-the-art methods, the proposed semi- and self-supervised models achieve competitive and superior deconvolution and fusion quality in the two-view and quad-view cases, respectively.
翻译:荧光显微镜的最近发展使得随着时间的推移能够捕捉活型生物体的高分辨率 3D 图像。为了能够图像甚至大型样本,多视光谱成像技术等技术在每个时间点记录不同方向,然后可以整合成单一的高质量体积。根据测量的点分布功能(PSF),分变和内容融合能够在很大程度上恢复成像过程期间不可避免的退化。经典多视分解和聚合方法主要使用迭代程序和内容平均。最近,革命神经网络(CNN)被应用到接近3D单视光谱显微镜上,但多视图案例有待研究。我们研究了基于CNN的多视图变异和聚合的两种合成数据集的功效,它们模拟胚胎,涉及两种或四种互补的三维观点。与典型的状态方法相比,拟议的半和自我监督模型在两视和四视案例中分别实现了竞争性和高级变异变和变质量。