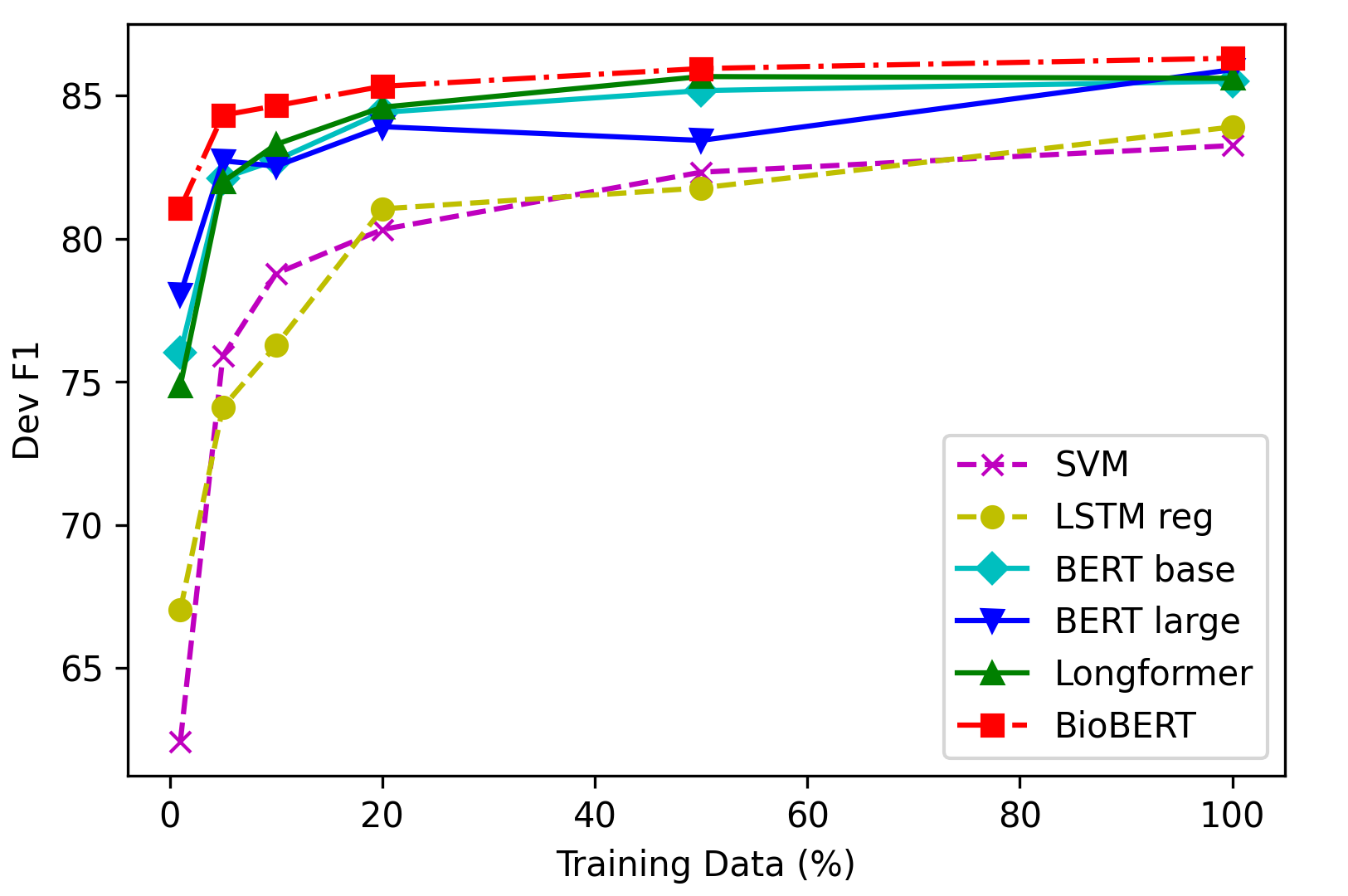
The global pandemic has made it more important than ever to quickly and accurately retrieve relevant scientific literature for effective consumption by researchers in a wide range of fields. We provide an analysis of several multi-label document classification models on the LitCovid dataset, a growing collection of 23,000 research papers regarding the novel 2019 coronavirus. We find that pre-trained language models fine-tuned on this dataset outperform all other baselines and that BioBERT surpasses the others by a small margin with micro-F1 and accuracy scores of around 86% and 75% respectively on the test set. We evaluate the data efficiency and generalizability of these models as essential features of any system prepared to deal with an urgent situation like the current health crisis. Finally, we explore 50 errors made by the best performing models on LitCovid documents and find that they often (1) correlate certain labels too closely together and (2) fail to focus on discriminative sections of the articles; both of which are important issues to address in future work. Both data and code are available on GitHub.
翻译:全球流行病使迅速和准确地检索相关科学文献,供研究人员在广泛领域有效使用比以往任何时候都更加重要。我们分析了LitCovid数据集的若干多标签文件分类模型,该数据集收集了23 000份关于新奇2019年科罗纳病毒的研究文件。我们发现,对这一数据进行微调的预先培训的语言模型比其他所有基线都高,生物生物-生物-生物-生物-生物-生物-生物-生物伦理学模型比其他模型高出很小的差幅,微型-F1和测试集的精确分分别为86%和75%左右。我们评估了这些模型的数据效率和可概括性,作为任何系统的基本特征,准备应对当前健康危机等紧急情况。最后,我们探索了在LitCovid文件上最优秀表现的模型造成的50个错误,发现这些错误往往(1) 将某些标签过于紧密地联系在一起,(2) 未能侧重于文章中的歧视性部分;两者都是今后工作中需要解决的重要问题。两种数据和代码都放在GitHub上。