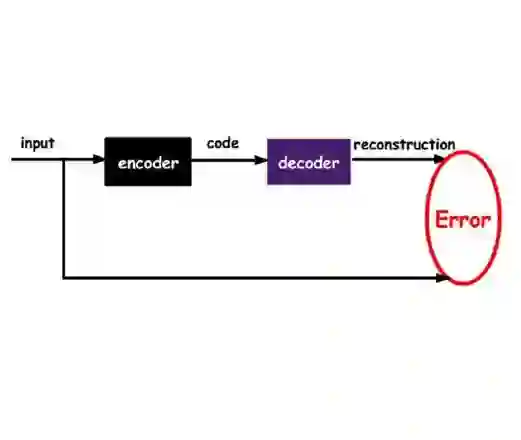
In recent years, deep generative models have attracted increasing interest due to their capacity to model complex distributions. Among those models, variational autoencoders have gained popularity as they have proven both to be computationally efficient and yield impressive results in multiple fields. Following this breakthrough, extensive research has been done in order to improve the original publication, resulting in a variety of different VAE models in response to different tasks. In this paper we present Pythae, a versatile open-source Python library providing both a unified implementation and a dedicated framework allowing straightforward, reproducible and reliable use of generative autoencoder models. We then propose to use this library to perform a case study benchmark where we present and compare 19 generative autoencoder models representative of some of the main improvements on downstream tasks such as image reconstruction, generation, classification, clustering and interpolation. The open-source library can be found at https://github.com/clementchadebec/benchmark_VAE.
翻译:近年来,深层基因模型因其模拟复杂分布的能力而引起越来越多的兴趣,在这些模型中,变异自动编码器越来越受欢迎,因为它们证明具有计算效率,在多个领域产生令人印象深刻的结果。在取得这一突破之后,为了改进原始出版物,进行了广泛的研究,从而根据不同的任务,产生了各种不同的VAE模型。在本文件中,我们介绍了Pythae,一个多用途的开放源Python图书馆,它提供了一个统一的实施和专用框架,允许直接、可复制和可靠地使用基因自动编码模型。我们然后提议利用这个图书馆进行案例研究基准,其中我们介绍并比较了19个基因自动编码模型,这些模型代表了下游任务的一些主要改进,例如图像重建、生成、分类、集群和内插。开放源图书馆见https://github.com/centchadebec/benchmart_VAE。