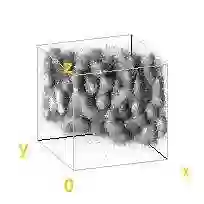
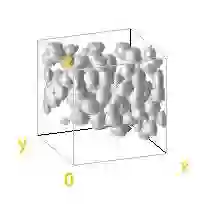
Automated image processing approaches are indispensable for many biomedical experiments and help to cope with the increasing amount of microscopy image data in a fast and reproducible way. Especially state-of-the-art deep learning-based approaches most often require large amounts of annotated training data to produce accurate and generalist outputs, but they are often compromised by the general lack of those annotated data sets. In this work, we propose how conditional generative adversarial networks can be utilized to generate realistic image data for 3D fluorescence microscopy from annotation masks of 3D cellular structures. In combination with mask simulation approaches, we demonstrate the generation of fully-annotated 3D microscopy data sets that we make publicly available for training or benchmarking. An additional positional conditioning of the cellular structures enables the reconstruction of position-dependent intensity characteristics and allows to generate image data of different quality levels. A patch-wise working principle and a subsequent full-size reassemble strategy is used to generate image data of arbitrary size and different organisms. We present this as a proof-of-concept for the automated generation of fully-annotated training data sets requiring only a minimum of manual interaction to alleviate the need of manual annotations.
翻译:自动化图像处理方法对于许多生物医学实验是不可或缺的,有助于以快速和可复制的方式处理越来越多的显微镜图像数据。特别是最先进的深层次学习方法,往往需要大量附加说明的培训数据,才能产生准确和一般性的产出,但往往由于普遍缺乏这些附加说明的数据集而受到影响。在这项工作中,我们提议如何利用有条件的基因化对抗网络,为3D 3D 细胞结构的注解面罩的3D 荧光显微镜生成现实的图像数据。我们结合面具模拟方法,展示了充分加注的3D 显微镜数据集的生成,我们公开提供用于培训或基准测试。细胞结构的附加定位性调整使得能够重建依赖位置的强度特性,并能够生成不同质量水平的图像数据。我们采用了一种不完全的工作原则和随后的全尺寸重新组合战略来生成任意尺寸和不同生物体的图像数据。我们将此作为自动生成充分附加说明的培训数据集的验证概念,只需要一个最低限度的手动互动来减轻对手册的需求。