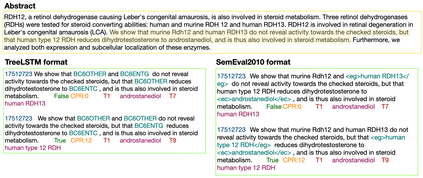
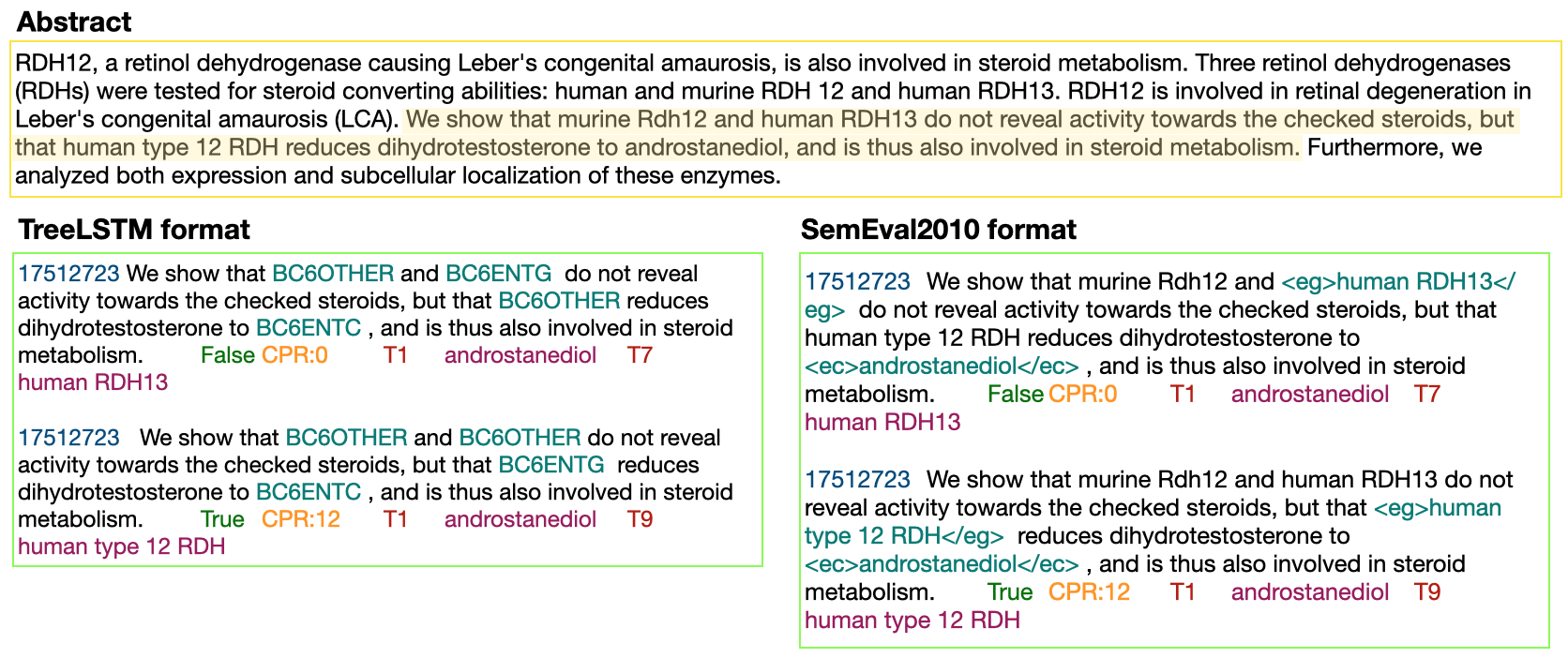
In Track-1 of the BioCreative VII Challenge participants are asked to identify interactions between drugs/chemicals and proteins. In-context named entity annotations for each drug/chemical and protein are provided and one of fourteen different interactions must be automatically predicted. For this relation extraction task, we attempt both a BERT-based sentence classification approach, and a more novel text-to-text approach using a T5 model. We find that larger BERT-based models perform better in general, with our BioMegatron-based model achieving the highest scores across all metrics, achieving 0.74 F1 score. Though our novel T5 text-to-text method did not perform as well as most of our BERT-based models, it outperformed those trained on similar data, showing promising results, achieving 0.65 F1 score. We believe a text-to-text approach to relation extraction has some competitive advantages and there is a lot of room for research advancement.
翻译:在第七轮《生物动力挑战》第1轨中,参与者被要求确定药物/化学和蛋白质之间的相互作用,提供每种药物/化学和蛋白质的内置名称实体说明,必须自动预测14种不同相互作用中的一个。对于这一关系提取任务,我们试图采用基于BERT的判刑分类方法,并采用采用T5模式的更新颖的文本对文本的方法。我们认为,基于BERT的更大模型总体上表现更好,我们基于BEERT的模型在所有指标中得分最高,达到0.74 F1分。尽管我们新型的T5文本对文本方法没有发挥以及我们基于BERT的大多数模型,但它优于那些受过类似数据培训的模型,显示有希望的结果,达到0.65 F1分。我们认为,采用文本对文本方法进行关联提取具有一些竞争优势,而且研究进展的空间很大。