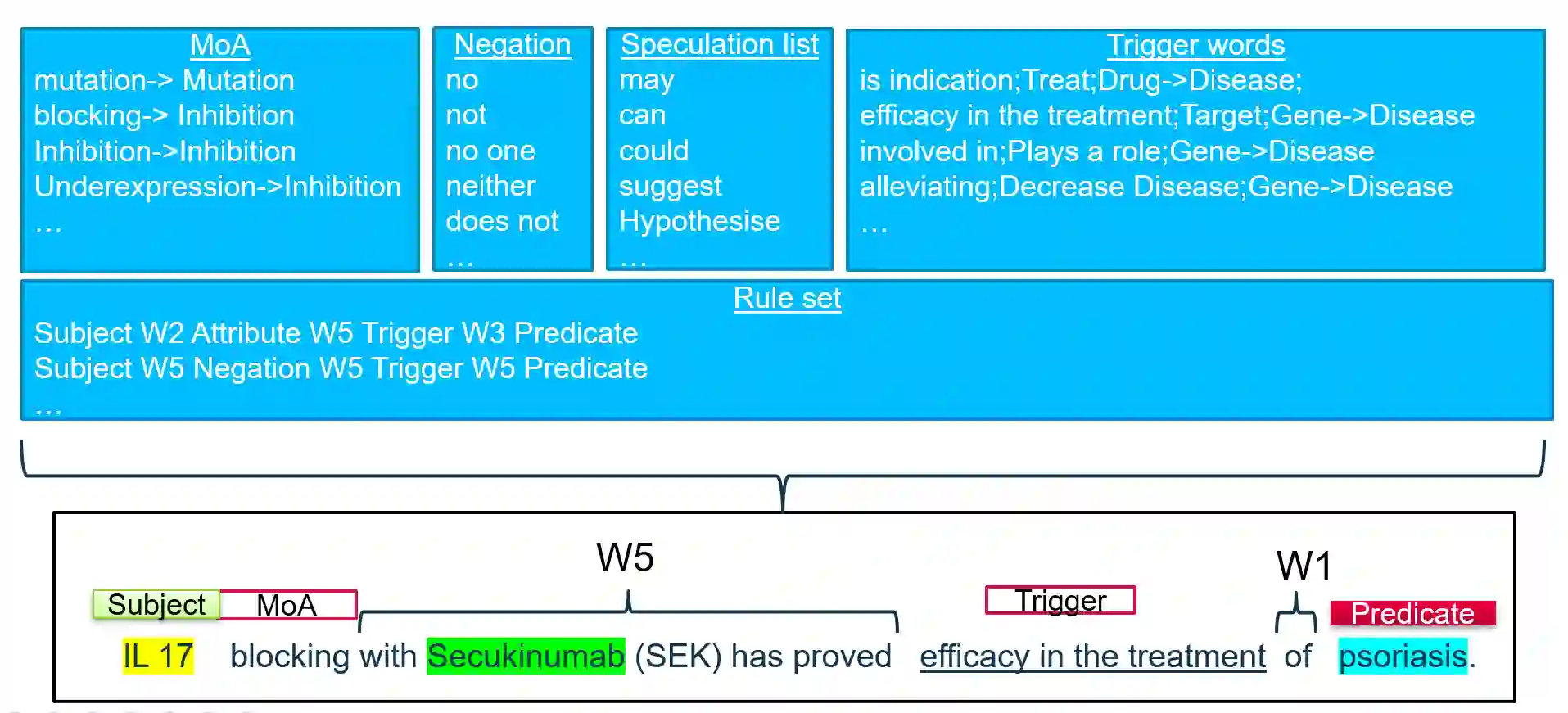
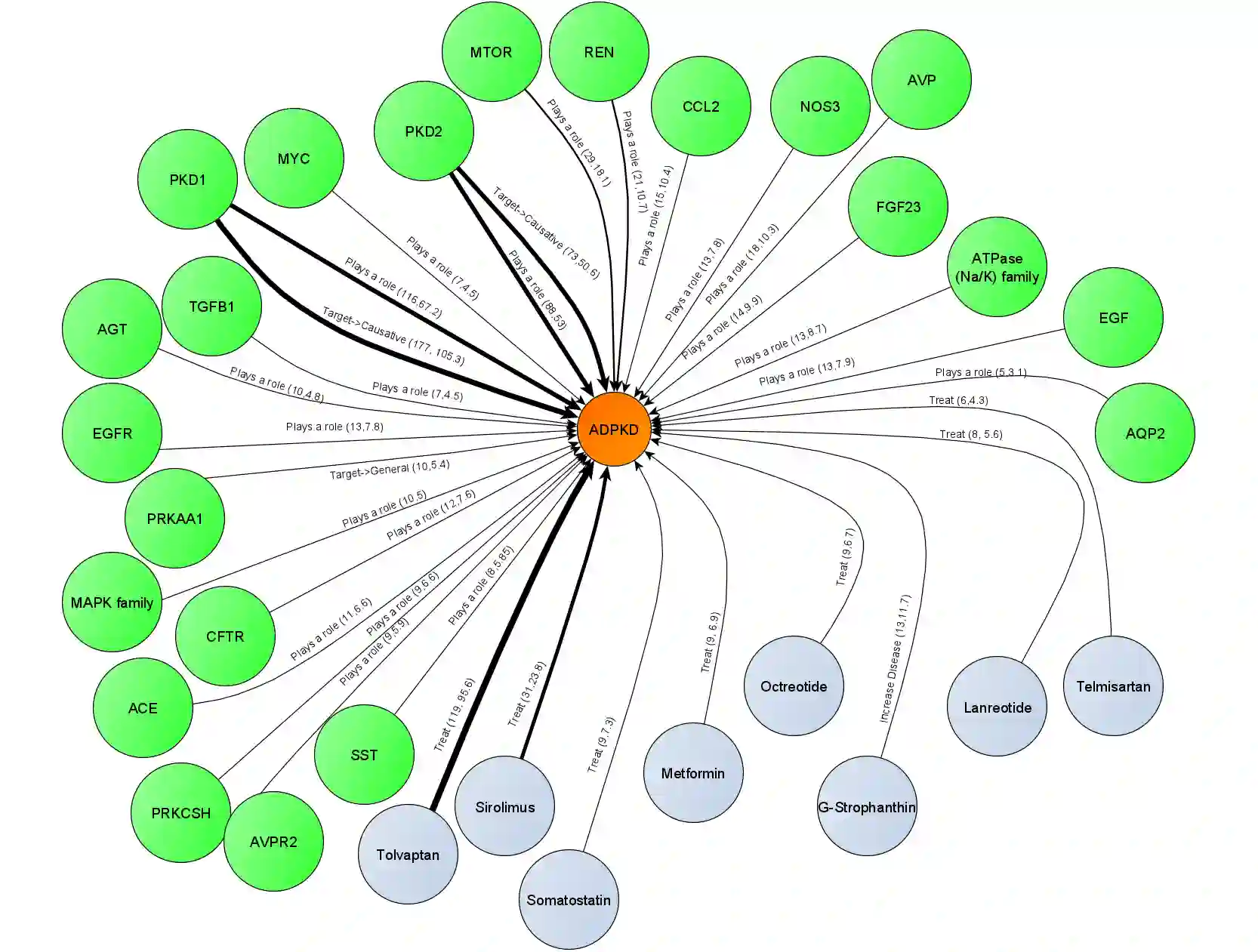
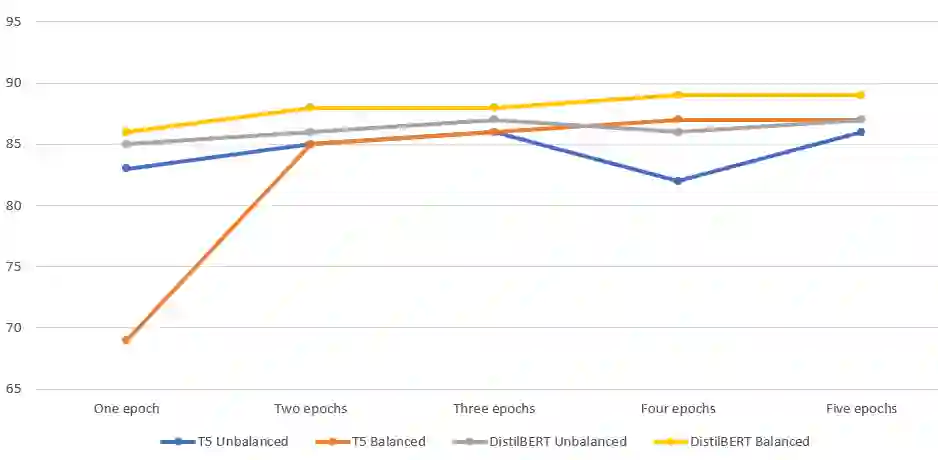
Biomedical research is growing at such an exponential pace that scientists, researchers, and practitioners are no more able to cope with the amount of published literature in the domain. The knowledge presented in the literature needs to be systematized in such a way that claims and hypotheses can be easily found, accessed, and validated. Knowledge graphs can provide such a framework for semantic knowledge representation from literature. However, in order to build a knowledge graph, it is necessary to extract knowledge as relationships between biomedical entities and normalize both entities and relationship types. In this paper, we present and compare a few rule-based and machine learning-based (Naive Bayes, Random Forests as examples of traditional machine learning methods and DistilBERT and T5-based models as examples of modern deep learning transformers) methods for scalable relationship extraction from biomedical literature, and for the integration into the knowledge graphs. We examine how resilient are these various methods to unbalanced and fairly small datasets. Our experiments show that transformer-based models handle well both small (due to pre-training on a large dataset) and unbalanced datasets. The best performing model was the DistilBERT-based model fine-tuned on balanced data, with a reported F1-score of 0.89.
翻译:生物医学研究正在以如此迅猛的速度发展,以至于科学家、研究人员和从业者无法更有能力应付这个领域大量出版的文献。文献中提供的知识需要系统化,以便能够容易地找到、获取和验证各种主张和假设。知识图表可以为文献中的语义知识的表达提供这样一个框架。然而,为了建立知识图,有必要提取知识,作为生物医学实体之间的关系,并使生物医学实体和实体及关系类型正常化。在本文中,我们介绍并比较少数基于规则和机器学习的少数基于规则的学习文献(Naive Bayes、随机森林作为传统机器学习方法的范例,DistillBERT和T5模型作为现代深层学习变异器的范例),以便从生物医学文献中提取可缩放关系的方法,以及将之纳入知识图中。我们研究了这些不同方法对不平衡和相当小的数据集的适应性。我们的实验表明,基于变异模型处理得既小(由于对大型数据集进行预先培训),也不平衡的数据集。最佳的模型是据报告的DastilBERT-BRET核心数据平衡模型。