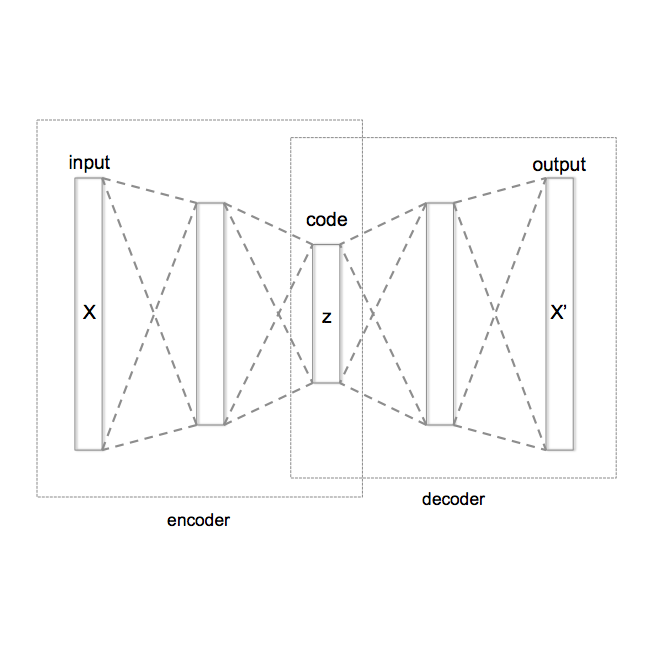
The chemical space of drug-like molecules is vast, motivating the development of generative models that must learn broad chemical distributions, enable conditional generation by capturing structure-property representations, and provide fast molecular generation. Meeting the objectives depends on modeling choices, including the probabilistic modeling approach, the conditional generative formulation, the architecture, and the molecular input representation. To address the challenges, we present STAR-VAE (Selfies-encoded, Transformer-based, AutoRegressive Variational Auto Encoder), a scalable latent-variable framework with a Transformer encoder and an autoregressive Transformer decoder. It is trained on 79 million drug-like molecules from PubChem, using SELFIES to guarantee syntactic validity. The latent-variable formulation enables conditional generation: a property predictor supplies a conditioning signal that is applied consistently to the latent prior, the inference network, and the decoder. Our contributions are: (i) a Transformer-based latent-variable encoder-decoder model trained on SELFIES representations; (ii) a principled conditional latent-variable formulation for property-guided generation; and (iii) efficient finetuning with low-rank adapters (LoRA) in both encoder and decoder, enabling fast adaptation with limited property and activity data. On the GuacaMol and MOSES benchmarks, our approach matches or exceeds baselines, and latent-space analyses reveal smooth, semantically structured representations that support both unconditional exploration and property-aware generation. On the Tartarus benchmarks, the conditional model shifts docking-score distributions toward stronger predicted binding. These results suggest that a modernized, scale-appropriate VAE remains competitive for molecular generation when paired with principled conditioning and parameter-efficient finetuning.
翻译:类药分子的化学空间极为广阔,这推动了生成模型的发展,这些模型需学习广泛的化学分布、通过捕获结构-性质表征实现条件生成,并提供快速的分子生成。实现这些目标取决于建模选择,包括概率建模方法、条件生成形式、架构以及分子输入表示。为应对这些挑战,我们提出了STAR-VAE(基于SELFIES编码的Transformer自回归变分自编码器),这是一个可扩展的隐变量框架,包含Transformer编码器和自回归Transformer解码器。该模型在PubChem的7900万个类药分子上进行训练,使用SELFIES表示保证语法有效性。隐变量形式支持条件生成:性质预测器提供条件信号,该信号被一致地应用于隐变量先验、推断网络和解码器。我们的贡献包括:(i)基于SELFIES表示训练的Transformer隐变量编码器-解码器模型;(ii)用于性质引导生成的原则性条件隐变量形式;(iii)在编码器和解码器中采用低秩适配器(LoRA)进行高效微调,实现在有限性质和活性数据下的快速适应。在GuacaMol和MOSES基准测试中,我们的方法达到或超越了基线模型,隐空间分析揭示了平滑且具有语义结构的表征,支持无条件探索和性质感知生成。在Tartarus基准测试中,条件模型将对接分数分布向预测结合力更强的方向偏移。这些结果表明,当结合原则性条件生成与参数高效微调时,现代化且规模适配的VAE在分子生成任务中仍具竞争力。