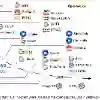
Foundation models in artificial intelligence (AI) are transforming medical imaging by enabling general-purpose feature learning from large-scale, unlabeled datasets. In this work, we introduce BrainFound, a self-supervised foundation model for brain MRI, built by extending DINO-v2, a vision transformer originally designed for 2D natural images. BrainFound adapts DINO-v2 to model full 3D brain anatomy by incorporating volumetric information from sequential MRI slices, moving beyond conventional single-slice paradigms. It supports both single- and multimodal inputs, enabling a broad range of downstream tasks, including disease detection and image segmentation, while generalising across varied imaging protocols and clinical scenarios. We show that BrainFound consistently outperforms existing self-supervised pretraining strategies and supervised baselines, particularly in label-scarce and multi-contrast settings. By integrating information from diverse 3D MRI modalities (e.g., T1, T2, FLAIR), it enhances diagnostic accuracy and reduces dependency on extensive expert annotations. This flexibility makes BrainFound a scalable and practical solution for 3D neuroimaging pipelines, with significant potential for clinical deployment and research innovation.
翻译:人工智能中的基础模型正通过从大规模无标注数据集中学习通用特征,推动医学影像领域的变革。本研究提出BrainFound——一种用于脑部磁共振成像的自监督基础模型,该模型通过扩展DINO-v2(一种最初为二维自然图像设计的视觉Transformer)构建而成。BrainFound通过整合连续MRI切片的体数据信息,将DINO-v2适配于全三维脑部解剖结构建模,突破了传统单切片范式的局限。该模型支持单模态与多模态输入,能够适应不同成像协议与临床场景,实现包括疾病检测与图像分割在内的广泛下游任务。实验表明,BrainFound在标签稀缺和多对比度场景下持续优于现有自监督预训练策略及有监督基线方法。通过融合多种三维MRI模态数据(如T1、T2、FLAIR),该模型提升了诊断准确性并降低了对大量专家标注的依赖。这种灵活性使BrainFound成为三维神经影像流程中可扩展的实用解决方案,在临床部署与科研创新方面具有显著潜力。