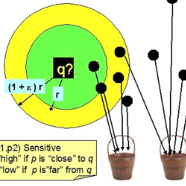
We present a new algorithm for the approximate near neighbor problem that combines classical ideas from group testing with locality-sensitive hashing (LSH). We reduce the near neighbor search problem to a group testing problem by designating neighbors as "positives," non-neighbors as "negatives," and approximate membership queries as group tests. We instantiate this framework using distance-sensitive Bloom Filters to Identify Near-Neighbor Groups (FLINNG). We prove that FLINNG has sub-linear query time and show that our algorithm comes with a variety of practical advantages. For example, FLINNG can be constructed in a single pass through the data, consists entirely of efficient integer operations, and does not require any distance computations. We conduct large-scale experiments on high-dimensional search tasks such as genome search, URL similarity search, and embedding search over the massive YFCC100M dataset. In our comparison with leading algorithms such as HNSW and FAISS, we find that FLINNG can provide up to a 10x query speedup with substantially smaller indexing time and memory.
翻译:我们为近邻的近邻问题提出了一个新的算法,它结合了从群体测试到对地点敏感的散列(LSH)的古典想法。我们通过将邻居指定为“阳性”,将非邻居指定为“负性”,将会员询问大致定为“负性”以及将会员问询作为集体测试。我们使用远程敏感的闪烁过滤器对这个框架进行即时应用,以识别近邻群体(FLINNG) 。我们证明FLINNG有亚线查询时间,并表明我们的算法具有各种实际优势。例如,通过数据,可以通过一个单一的传票构建FLINNG,完全由有效的整数操作组成,不需要任何距离计算。我们在基因组搜索、URm相似性搜索和嵌入大型 YFCC100M 数据集中进行大规模搜索等高度的实验。我们与HNSW和FISS相比,我们发现FLINNG可以提供10x查询速度,而索引时间和记忆要小得多。