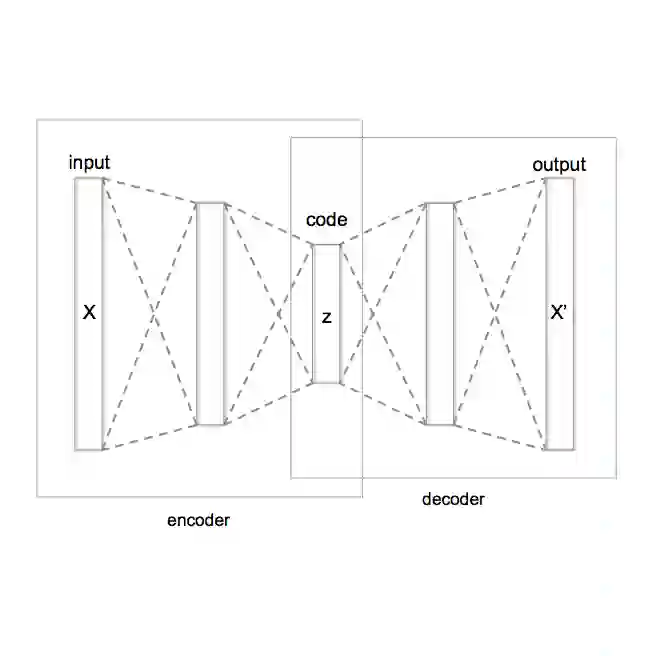
Single-cell RNA sequencing (scRNA-seq) has revealed complex cellular heterogeneity, but recent studies emphasize that understanding biological function also requires modeling cell-cell communication (CCC), the signaling interactions mediated by ligand-receptor pairs that coordinate cellular behavior. Tools like CellChat have demonstrated that CCC plays a critical role in processes such as cell differentiation, tissue regeneration, and immune response, and that transcriptomic data inherently encodes rich information about intercellular signaling. We propose CCCVAE, a novel variational autoencoder framework that incorporates CCC signals into single-cell representation learning. By leveraging a communication-aware kernel derived from ligand-receptor interactions and a sparse Gaussian process, CCCVAE encodes biologically informed priors into the latent space. Unlike conventional VAEs that treat each cell independently, CCCVAE encourages latent embeddings to reflect both transcriptional similarity and intercellular signaling context. Empirical results across four scRNA-seq datasets show that CCCVAE improves clustering performance, achieving higher evaluation scores than standard VAE baselines. This work demonstrates the value of embedding biological priors into deep generative models for unsupervised single-cell analysis.
翻译:单细胞RNA测序(scRNA-seq)技术揭示了复杂的细胞异质性,但近期研究强调,理解生物学功能还需对细胞间通信(CCC)进行建模——即由配体-受体对介导、协调细胞行为的信号交互作用。CellChat等工具已证明CCC在细胞分化、组织再生和免疫应答等过程中发挥关键作用,且转录组数据本身编码着丰富的细胞间信号传递信息。本文提出CCCVAE,一种将CCC信号整合到单细胞表征学习中的新型变分自编码器框架。通过利用源自配体-受体相互作用的通信感知核与稀疏高斯过程,CCCVAE将生物学先验信息编码到隐空间中。与将每个细胞独立处理的传统VAE不同,CCCVAE促使隐嵌入同时反映转录相似性与细胞间信号传导背景。在四个scRNA-seq数据集上的实验结果表明,CCCVAE提升了聚类性能,其评估分数优于标准VAE基线。本工作证明了将生物学先验嵌入深度生成模型对于无监督单细胞分析的价值。