入门必备!生物医学命名实体识别(BioNER)最全论文清单,附SOTA结果汇总
作者丨罗凌
学校丨大连理工大学博士
研究方向丨深度学习、文本分类
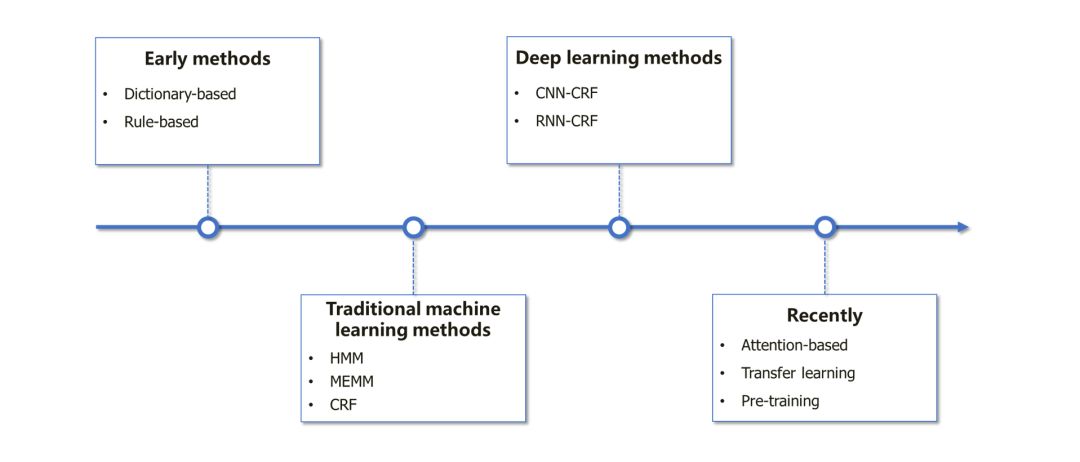
▶▷ 综述论文
Overview of BioCreative II gene mention recognition. Smith L, Tanabe L K, nee Ando R J, et al. Genome biology, 2008, 9(2): S2.
Biomedical named entity recognition: a survey of machine-learning tools. Campos D, Matos S, Oliveira J L. Theory and Applications for Advanced Text Mining, 2012: 175-195.
Chemical named entities recognition: a review on approaches and applications. Eltyeb S, Salim N. Journal of cheminformatics, 2014, 6(1): 17.
CHEMDNER: The drugs and chemical names extraction challenge. Krallinger M, Leitner F, Rabal O, et al. Journal of cheminformatics, 2015, 7(1): S1.
A comparative study for biomedical named entity recognition. Wang X, Yang C, Guan R. International Journal of Machine Learning and Cybernetics, 2015, 9(3): 373-382.
▷▶ 基于词典的方法
Using BLAST for identifying gene and protein names in journal articles. Krauthammer M, Rzhetsky A, Morozov P, et al. Gene, 2000, 259(1-2): 245-252.
Boosting precision and recall of dictionary-based protein name recognition. Tsuruoka Y, Tsujii J. Proceedings of the ACL 2003 workshop on Natural language processing in biomedicine-Volume 13, 2003: 41-48.
Exploiting the performance of dictionary-based bio-entity name recognition in biomedical literature. Yang Z, Lin H, Li Y. Computational Biology and Chemistry, 2008, 32(4): 287-291.
A dictionary to identify small molecules and drugs in free text. Hettne K M, Stierum R H, Schuemie M J, et al. Bioinformatics, 2009, 25(22): 2983-2991.
LINNAEUS: a species name identification system for biomedical literature. Gerner M, Nenadic G, Bergman C M. BMC bioinformatics, 2010, 11(1): 85.
▷▶ 基于规则的方法
Toward information extraction: identifying protein names from biological papers. Fukuda K, Tsunoda T, Tamura A, et al. Pac symp biocomput. 1998, 707(18): 707-718.
A biological named entity recognizer. Narayanaswamy M, Ravikumar K E, Vijay-Shanker K. Biocomputing 2003. 2002: 427-438.
ProMiner: rule-based protein and gene entity recognition. Hanisch D, Fundel K, Mevissen H T, et al. BMC bioinformatics, 2005, 6(1): S14.
MutationFinder: a high-performance system for extracting point mutation mentions from text. Caporaso J G, Baumgartner Jr W A, Randolph D A, et al. Bioinformatics, 2007, 23(14): 1862-1865.
Drug name recognition and classification in biomedical texts: a case study outlining approaches underpinning automated systems. Segura-Bedmar I, Martínez P, Segura-Bedmar M. Drug discovery today, 2008, 13(17-18): 816-823.
Investigation of unsupervised pattern learning techniques for bootstrap construction of a medical treatment lexicon. Xu R, Morgan A, Das A K, et al. Proceedings of the workshop on current trends in biomedical natural language processing, 2009: 63-70.
Linguistic approach for identification of medication names and related information in clinical narratives. Hamon T, Grabar N. Journal of the American Medical Informatics Association, 2010, 17(5): 549-554.
SETH detects and normalizes genetic variants in text. Thomas P, Rocktäschel T, Hakenberg J, et al. Bioinformatics, 2016, 32(18): 2883-2885.
PENNER: Pattern-enhanced Nested Named Entity Recognition in Biomedical Literature. Wang X, Zhang Y, Li Q, et al. 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2018: 540-547.
SVM-based Methods
Tuning support vector machines for biomedical named entity recognition. Kazama J, Makino T, Ohta Y, et al. Proceedings of the ACL-02 workshop on Natural language processing in the biomedical domain-Volume 3, 2002: 1-8.
Biomedical named entity recognition using two-phase model based on SVMs. Lee K J, Hwang Y S, Kim S, et al. Journal of Biomedical Informatics, 2004, 37(6): 436-447.
Exploring deep knowledge resources in biomedical name recognition. GuoDong Z, Jian S. Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications, 2004: 96-99.
Named entity recognition in biomedical texts using an HMM model. Zhao S. Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications, 2004: 84-87.
Annotation of chemical named entities. Corbett P, Batchelor C, Teufel S. Proceedings of the Workshop on BioNLP 2007: Biological, Translational, and Clinical Language Processing, 2007: 57-64.
Conditional random fields vs. hidden markov models in a biomedical named entity recognition task. Ponomareva N, Rosso P, Pla F, et al. Proc. of Int. Conf. Recent Advances in Natural Language Processing, RANLP. 2007, 479: 483.
Cascaded classifiers for confidence-based chemical named entity recognition. Corbett P, Copestake A. BMC bioinformatics, 2008, 9(11): S4.
OSCAR4: a flexible architecture for chemical text-mining. Jessop D M, Adams S E, Willighagen E L, et al. Journal of cheminformatics, 2011, 3(1): 41.
ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Settles B. Bioinformatics, 2005, 21(14): 3191-3192.
BANNER: an executable survey of advances in biomedical named entity recognition. Leaman R, Gonzalez G. Biocomputing 2008. 2008: 652-663.
https://psb.stanford.edu/psb-online/proceedings/psb08/leaman.pdf
Incorporating rich background knowledge for gene named entity classification and recognition. Li Y, Lin H, Yang Z. BMC bioinformatics, 2009, 10(1): 223.
A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. Jiang M, Chen Y, Liu M, et al. Journal of the American Medical Informatics Association, 2011, 18(5): 601-606.
ChemSpot: a hybrid system for chemical named entity recognition. Rocktäschel T, Weidlich M, Leser U. Bioinformatics, 2012, 28(12): 1633-1640.
Gimli: open source and high-performance biomedical name recognition. Campos D, Matos S, Oliveira J L. BMC bioinformatics, 2013, 14(1): 54.
tmVar: a text mining approach for extracting sequence variants in biomedical literature. Wei C H, Harris B R, Kao H Y, et al. Bioinformatics, 2013, 29(11): 1433-1439.
Evaluating word representation features in biomedical named entity recognition tasks. Tang B, Cao H, Wang X, et al. BioMed research international, 2014, 2014.
Drug name recognition in biomedical texts: a machine-learning-based method. He L, Yang Z, Lin H, et al. Drug discovery today, 2014, 19(5): 610-617.
tmChem: a high performance approach for chemical named entity recognition and normalization. Leaman R, Wei C H, Lu Z. Journal of cheminformatics, 2015, 7(1): S3.
GNormPlus: an integrative approach for tagging genes, gene families, and protein domains. Wei C H, Kao H Y, Lu Z. BioMed research international, 2015, 2015.
Mining chemical patents with an ensemble of open systems[J]. Leaman R, Wei C H, Zou C, et al. Database, 2016, 2016.
nala: text mining natural language mutation mentions. Cejuela J M, Bojchevski A, Uhlig C, et al. Bioinformatics, 2017, 33(12): 1852-1858.
Recurrent neural network models for disease name recognition using domain invariant features. Sahu S, Anand A. Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics. 2016: 2216-2225.
Deep learning with word embeddings improves biomedical named entity recognition. Habibi M, Weber L, Neves M, et al. Bioinformatics, 2017, 33(14): i37-i48.
A neural joint model for entity and relation extraction from biomedical text. Li F, Zhang M, Fu G, et al. BMC bioinformatics, 2017, 18(1): 198.
A neural network multi-task learning approach to biomedical named entity recognition. Crichton G, Pyysalo S, Chiu B, et al. BMC bioinformatics, 2017, 18(1): 368.
Disease named entity recognition from biomedical literature using a novel convolutional neural network. Zhao Z, Yang Z, Luo L, et al. BMC medical genomics, 2017, 10(5): 73.
An attention-based BiLSTM-CRF approach to document-level chemical named entity recognition. Luo L, Yang Z, Yang P, et al. Bioinformatics, 2018, 34(8): 1381-1388.
GRAM-CNN: a deep learning approach with local context for named entity recognition in biomedical text. Zhu Q, Li X, Conesa A, et al. Bioinformatics, 2018, 34(9): 1547-1554.
https://academic.oup.com/bioinformatics/article-abstract/34/9/1547/4764002
D3NER: biomedical named entity recognition using CRF-biLSTM improved with fine-tuned embeddings of various linguistic information. Dang T H, Le H Q, Nguyen T M, et al. Bioinformatics, 2018, 34(20): 3539-3546.
Transfer learning for biomedical named entity recognition with neural networks. Giorgi J M, Bader G D. Bioinformatics, 2018, 34(23): 4087-4094.
Label-Aware Double Transfer Learning for Cross-Specialty Medical Named Entity Recognition. Wang Z, Qu Y, Chen L, et al. NAACL. 2018: 1-15.
Recognizing irregular entities in biomedical text via deep neural networks. Li F, Zhang M, Tian B, et al. Pattern Recognition Letters, 2018, 105: 105-113.
Cross-type biomedical named entity recognition with deep multi-task learning. Wang X, Zhang Y, Ren X, et al. Bioinformatics, 2019, 35(10): 1745-1752.
Improving Chemical Named Entity Recognition in Patents with Contextualized Word Embeddings. Zhai Z, Nguyen D Q, Akhondi S, et al. Proceedings of the 18th BioNLP Workshop and Shared Task. 2019: 328-338.
Chinese Clinical Named Entity Recognition Using Residual Dilated Convolutional Neural Network with Conditional Random Field. Qiu J, Zhou Y, Wang Q, et al. IEEE Transactions on NanoBioscience, 2019, 18(3): 306-315.
A Neural Multi-Task Learning Framework to Jointly Model Medical Named Entity Recognition and Normalization. Zhao S, Liu T, Zhao S, et al. Proceedings of the AAAI Conference on Artificial Intelligence. 2019, 33: 817-824.
CollaboNet: collaboration of deep neural networks for biomedical named entity recognition. Yoon W, So C H, Lee J, et al. BMC bioinformatics, 2019, 20(10): 249.
BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Lee J, Yoon W, Kim S, et al. Bioinformatics, Advance article, 2019.
https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btz682/5566506
https://github.com/dmis-lab/biobert
https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/btz528/5523847?redirectedFrom=fulltext
TaggerOne: joint named entity recognition and normalization with semi-Markov Models. Leaman R, Lu Z. Bioinformatics, 2016, 32(18): 2839-2846.
A transition-based joint model for disease named entity recognition and normalization. Lou Y, Zhang Y, Qian T, et al. Bioinformatics, 2017, 33(15): 2363-2371.
https://academic.oup.com/bioinformatics/article-abstract/33/15/2363/3089942
https://github.com/louyinxia/jointRN
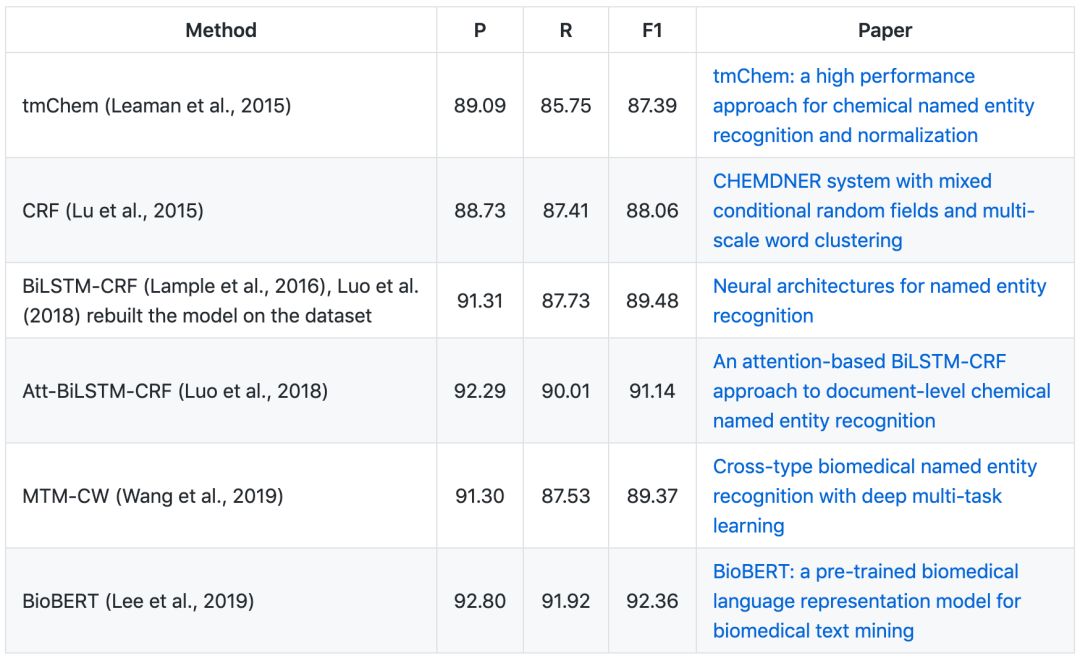
▶▷ Chemical NER
CHEMDNER
CHEMDNER (chemical compound and drug name recognition) task as part of the BioCreative IV challenge aims to promote the development of systems for the automatic recognition of chemical entities in text. It was divided into two tasks: one covered the indexing of documents with chemicals (chemical document indexing - CDI task), and the other was concerned with finding the exact mentions of chemicals in text (chemical entity mention recognition - CEM task). Here, we only focus on the CEM task.
The CHEMDNER corpus consists of 10,000 PubMed abstracts, which contains a total of 84,355 chemical entity mentions. The original corpus is divided into training set (3,500 abstracts), development set (3,500 abstracts) and test set (3,000 abstracts).
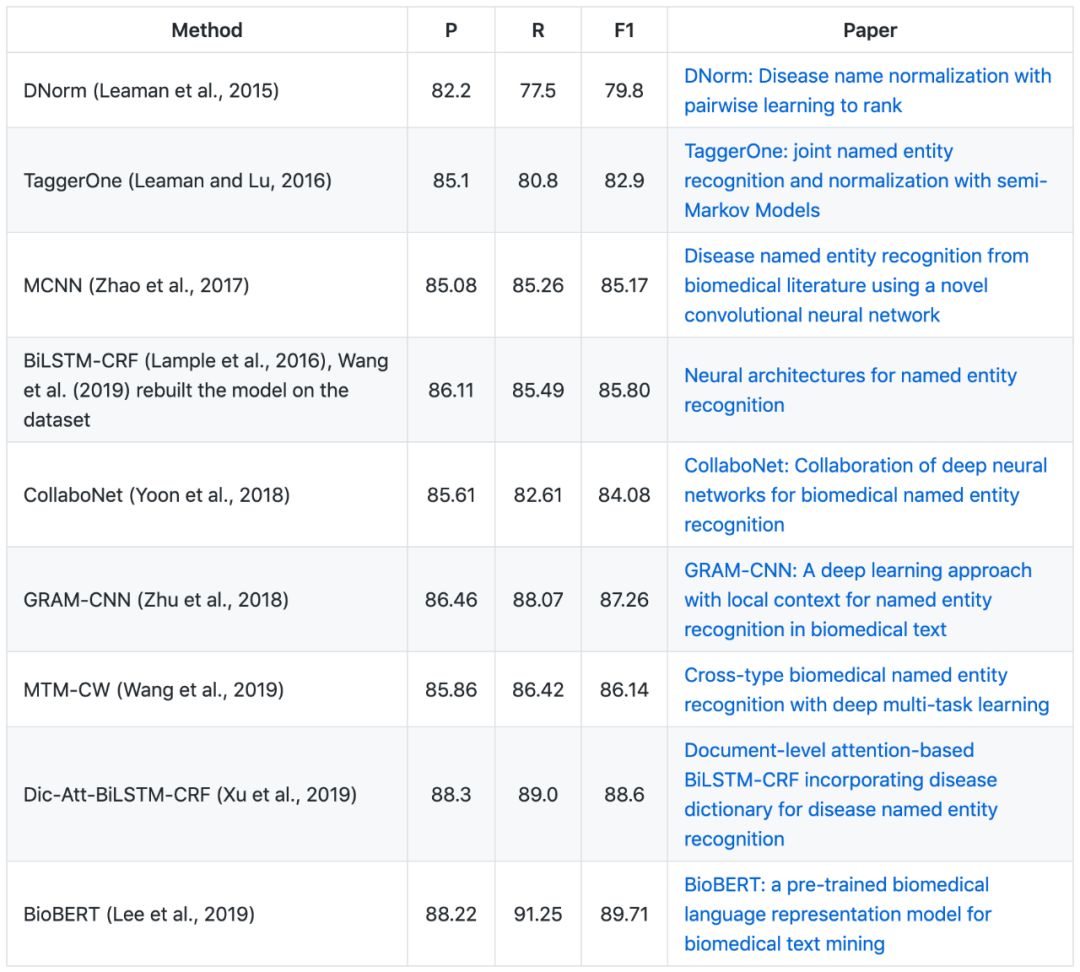
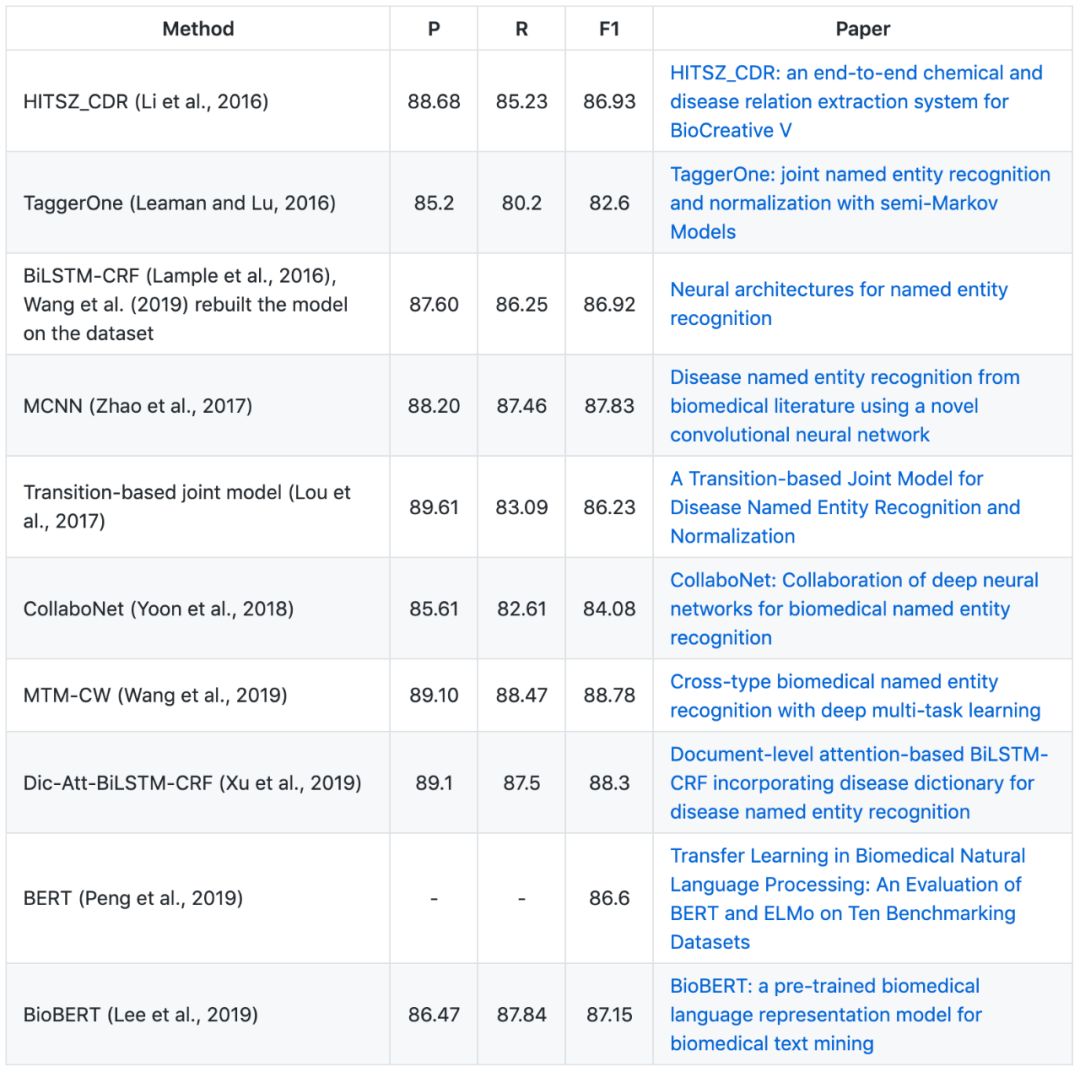
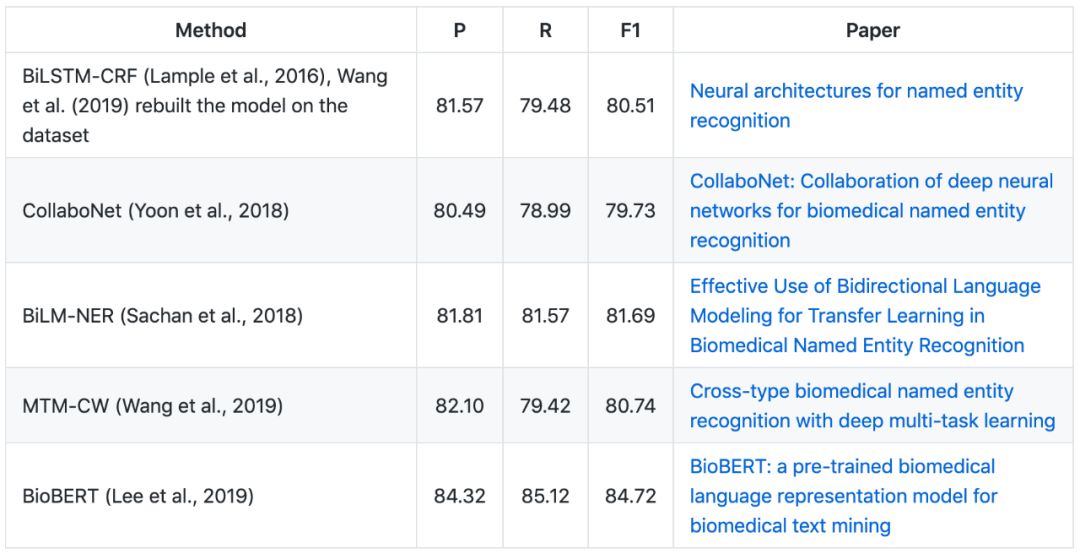
▶▷ Disease NER
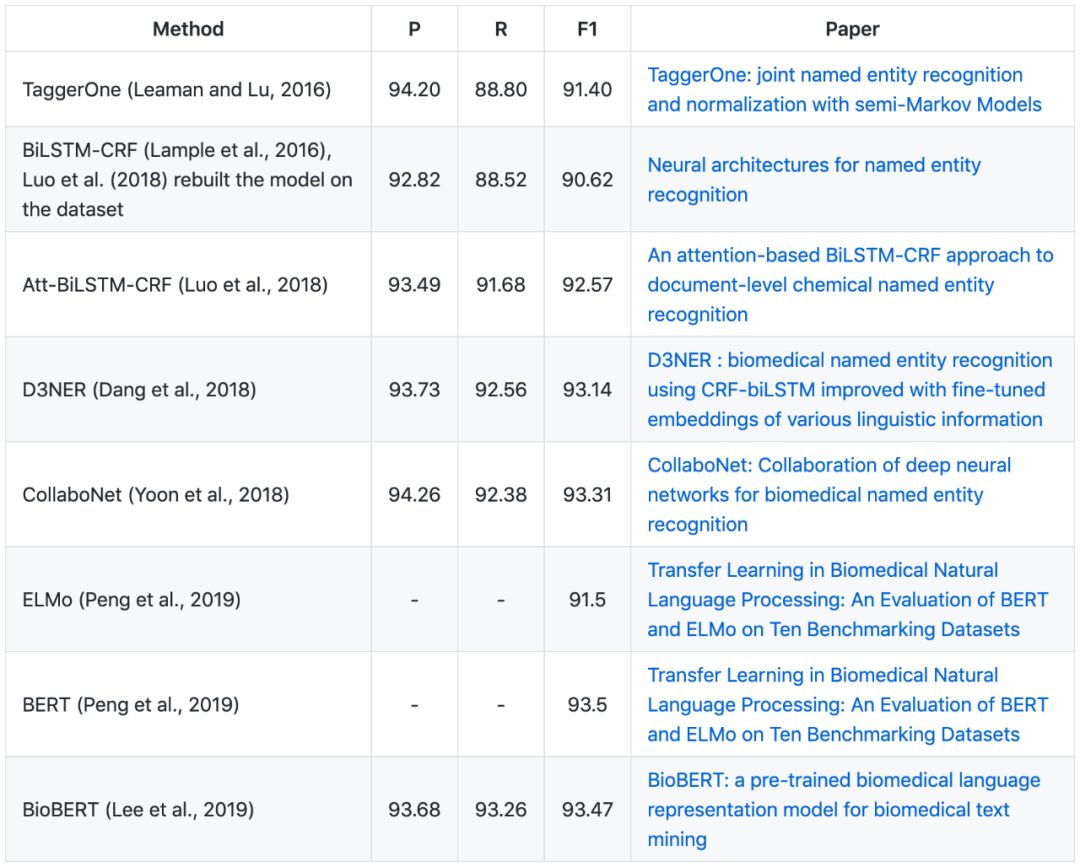
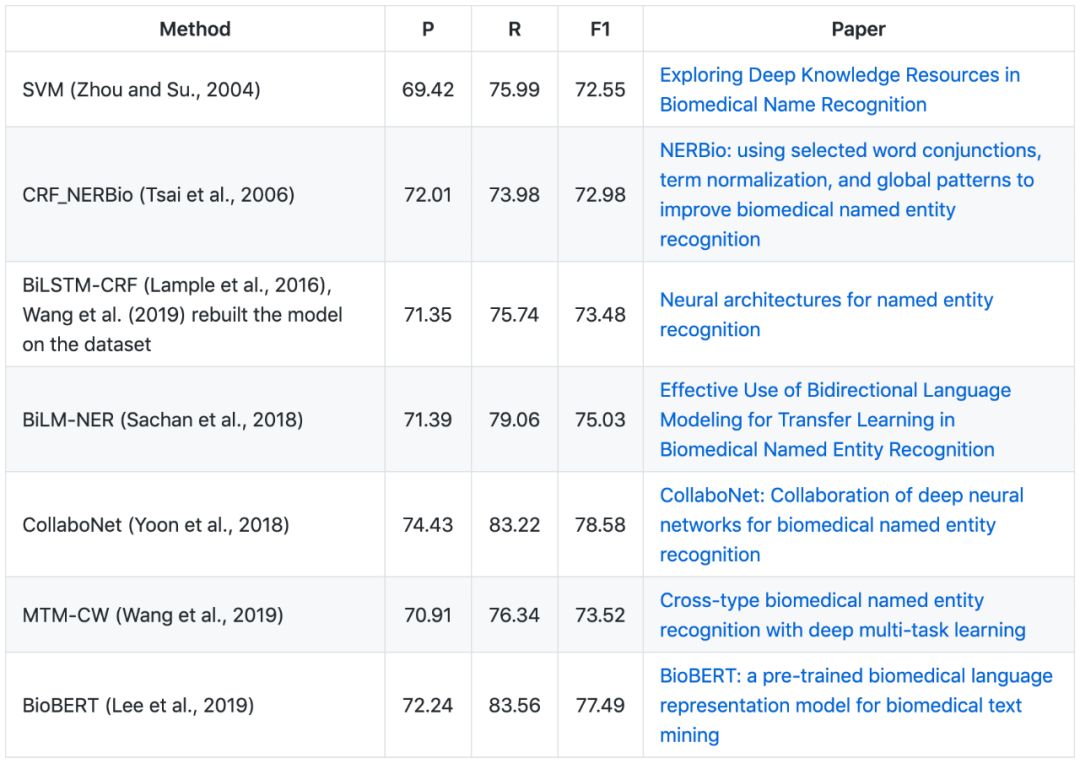
▶▷ Gene/Protein NER
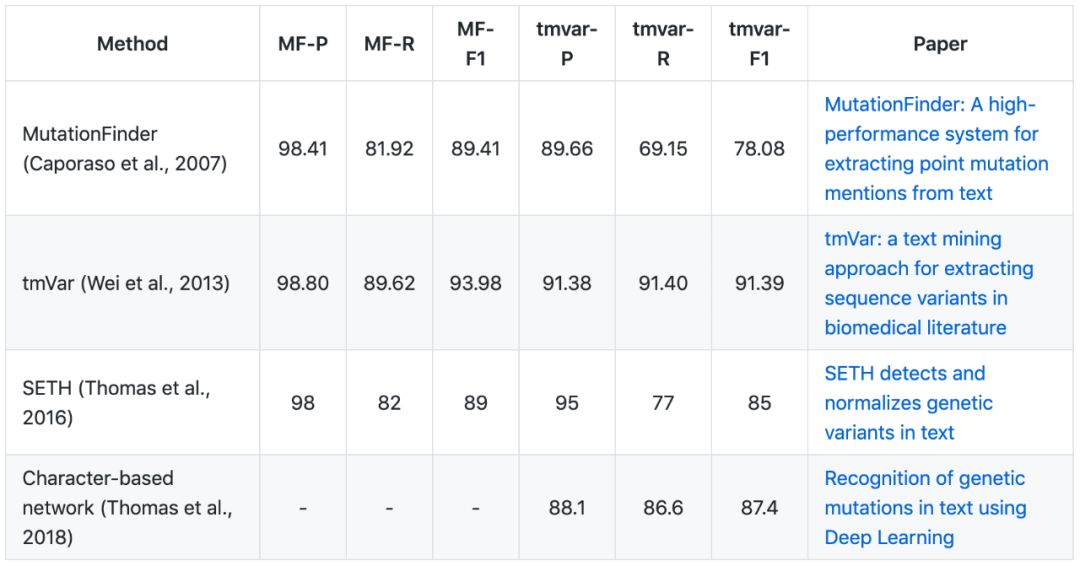
▶▷ Mutation NER
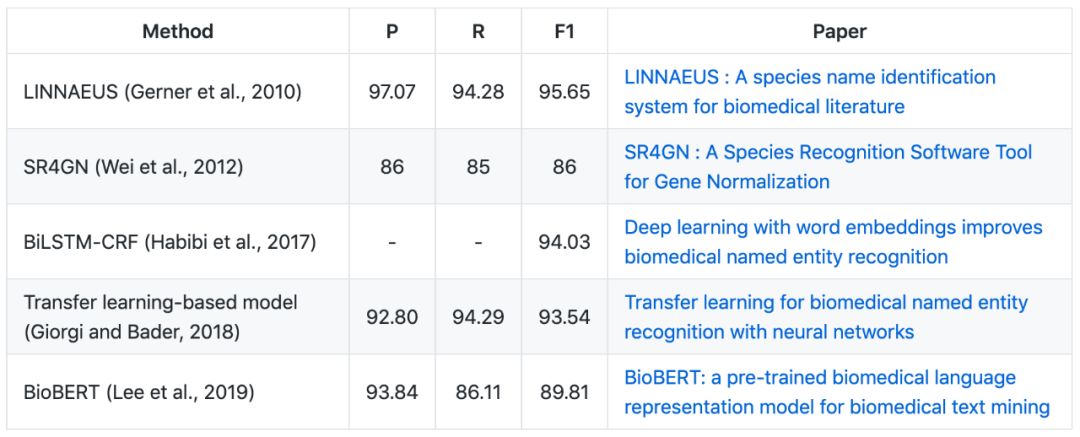
▶▷ Species NER
LINNAEUS corpus
The LINNAEUS corpus: A set of open access documents in text format, manually annotated for species mention tags. It consists of 100 full-text documents from the PMC OA document, which contains a total of 4,259 species entity mentions.
🔍
现在,在「知乎」也能找到我们了
进入知乎首页搜索「PaperWeekly」
点击「关注」订阅我们的专栏吧
关于PaperWeekly
PaperWeekly 是一个推荐、解读、讨论、报道人工智能前沿论文成果的学术平台。如果你研究或从事 AI 领域,欢迎在公众号后台点击「交流群」,小助手将把你带入 PaperWeekly 的交流群里。
▽ 点击 | 阅读原文 | 收藏论文清单