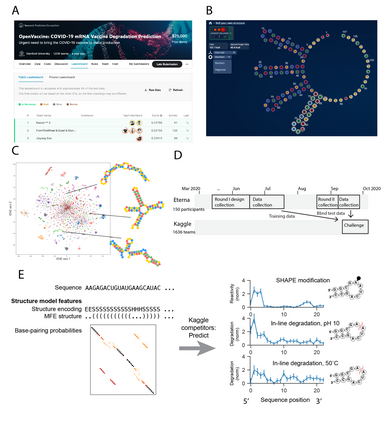
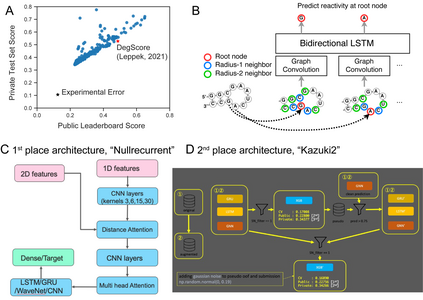
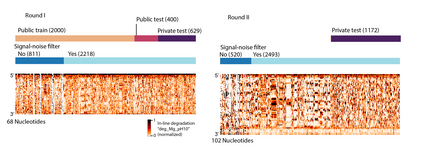
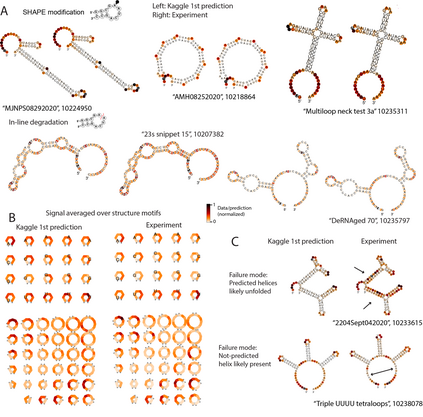
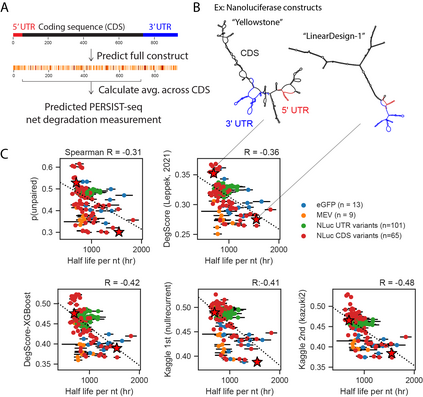
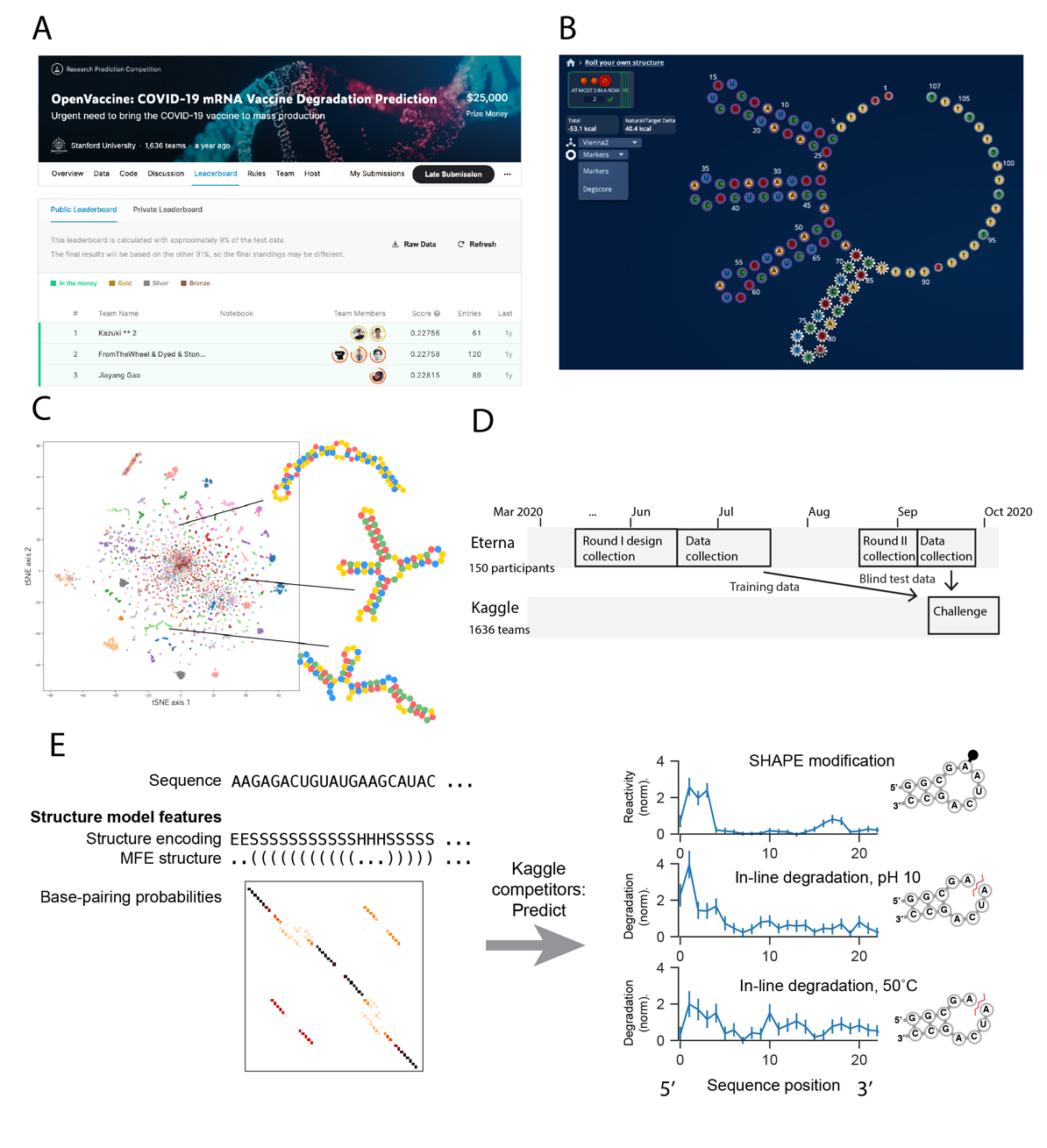
Messenger RNA-based medicines hold immense potential, as evidenced by their rapid deployment as COVID-19 vaccines. However, worldwide distribution of mRNA molecules has been limited by their thermostability, which is fundamentally limited by the intrinsic instability of RNA molecules to a chemical degradation reaction called in-line hydrolysis. Predicting the degradation of an RNA molecule is a key task in designing more stable RNA-based therapeutics. Here, we describe a crowdsourced machine learning competition ("Stanford OpenVaccine") on Kaggle, involving single-nucleotide resolution measurements on 6043 102-130-nucleotide diverse RNA constructs that were themselves solicited through crowdsourcing on the RNA design platform Eterna. The entire experiment was completed in less than 6 months. Winning models demonstrated test set errors that were better by 50% than the previous state-of-the-art DegScore model. Furthermore, these models generalized to blindly predicting orthogonal degradation data on much longer mRNA molecules (504-1588 nucleotides) with improved accuracy over DegScore and other models. Top teams integrated natural language processing architectures and data augmentation techniques with predictions from previous dynamic programming models for RNA secondary structure. These results indicate that such models are capable of representing in-line hydrolysis with excellent accuracy, supporting their use for designing stabilized messenger RNAs. The integration of two crowdsourcing platforms, one for data set creation and another for machine learning, may be fruitful for other urgent problems that demand scientific discovery on rapid timescales.
翻译:以RNA为主的药物具有巨大的潜力,正如它们作为COVID-19疫苗的迅速部署所证明的那样。然而,MRNA分子的可移动性限制了其在全世界的分布,因为分子的可移动性有限,而这种可移动性从根本上受到RNA分子内在的不稳定性的限制,成为称为线内水解化的化学降解反应。预测RNA分子的退化是设计更稳定的RNA治疗方法的关键任务。这里,我们描述了在Kaggle上由多方组成的机器学习竞赛(“斯坦福 OpenVaccine ”),其中包括对6043 102-130-Nucleotide多样化的RNA分子进行单核酸溶解度测量,这些分子本身通过RNA设计平台Eterna的众包收集。整个实验是在不到6个月的时间里完成的。 赢取模型显示的测试设置错误比以前最稳定的RNA治疗模型要好50%。 这些模型用于盲目地预测 mRNA分子(50-1588-108-NA)的直径流化数据(504-158 Nnucleotideal ) 。这些模型可以用来以更精确的快速的快速的流化数据模型,这些模型用来设计一个快速的精确的精确的流化模型,这些模型,用来用来模拟的流化数据结构的精确性模型,这些模型是用来在以前的流化数据结构的精确的精确化模型,这些模型,这些模型的精确性模型,用来用来用来用来用来用来用来用来进行精确化数据结构。