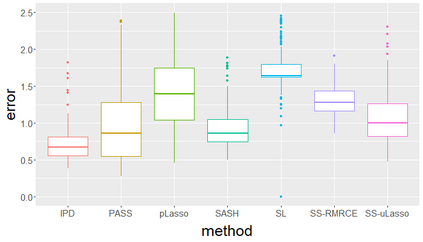
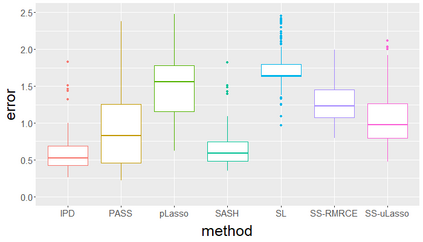
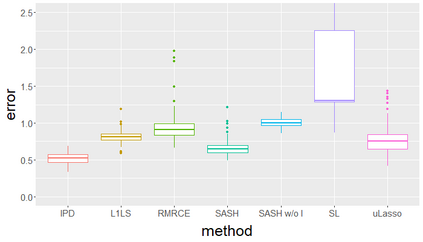
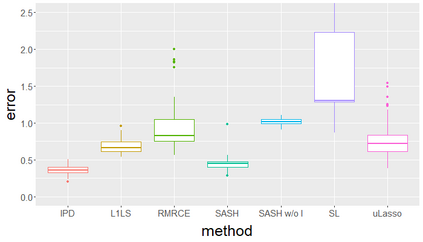
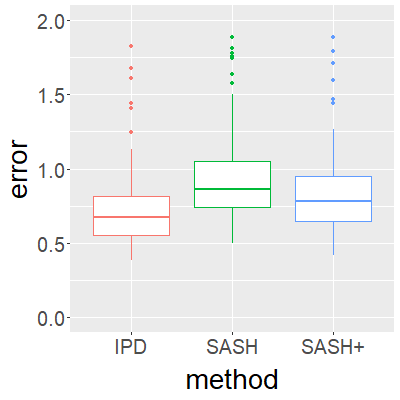
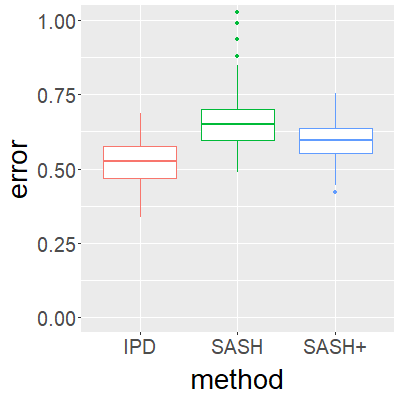
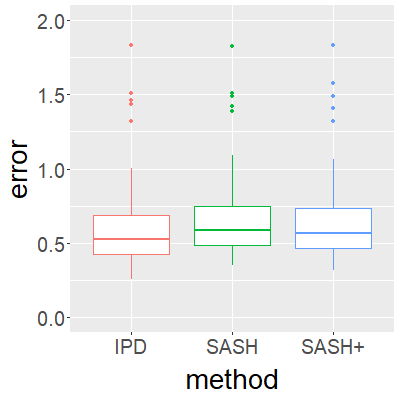
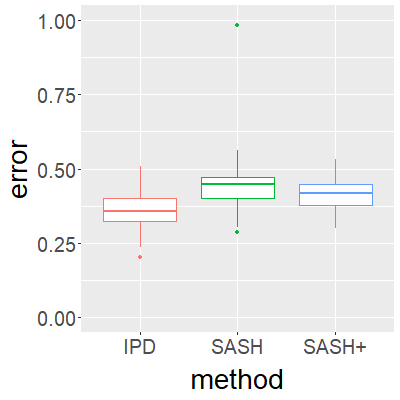
Surrogate variables in electronic health records (EHR) and biobank data play an important role in biomedical studies due to the scarcity or absence of chart-reviewed gold standard labels. We develop a novel approach named SASH for {\bf S}urrogate-{\bf A}ssisted and data-{\bf S}hielding {\bf H}igh-dimensional integrative regression. It is a semi-supervised approach that efficiently leverages sizable unlabeled samples with error-prone EHR surrogate outcomes from multiple local sites, to improve the learning accuracy of the small gold-labeled data. {To facilitate stable and efficient knowledge extraction from the surrogates, our method first obtains a preliminary supervised estimator, and then uses it to assist training a regularized single index model (SIM) for the surrogates. Interestingly, through a chain of convex and properly penalized sparse regressions that approximate the SIM loss with bias-correction, our method avoids the local minima issue of the SIM training, and fully eliminates the impact of the preliminary estimator's large error. In addition, it protects individual-level information through summary-statistics-based data aggregation across the local sites, leveraging a similar idea of bias-corrected approximation for SIM.} Through simulation studies, we demonstrate that our method outperforms existing approaches on finite samples. Finally, we apply our method to develop a high dimensional genetic risk model for type II diabetes using large-scale data sets from UK and Mass General Brigham biobanks, where only a small fraction of subjects in one site has been labeled via chart reviewing.
翻译:暂无翻译