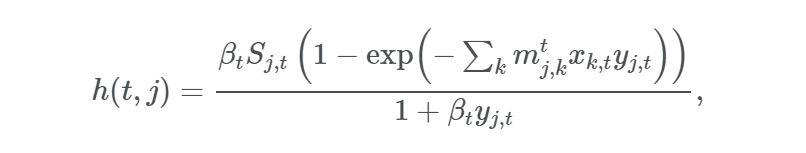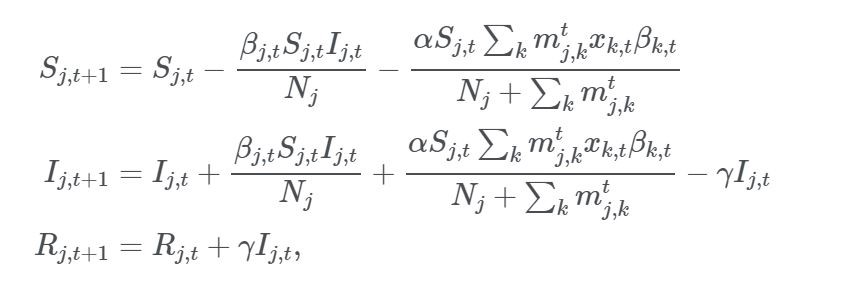疫情之下,这是你也能上手的Python新冠病毒传播建模教程(附代码)
选自 towardsdatascience
作者:Gevorg Yeghikyan
机器之心编译
机器之心编辑部
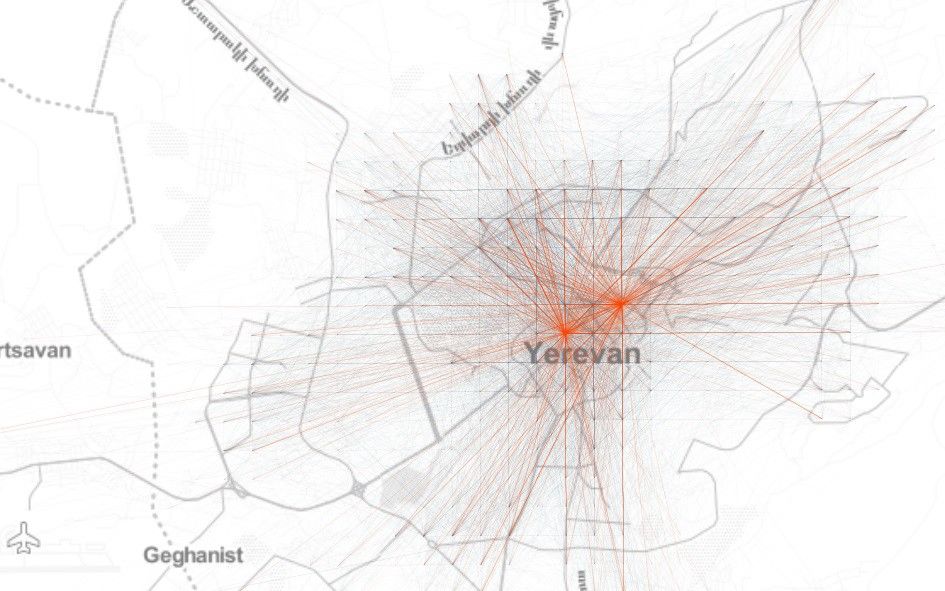
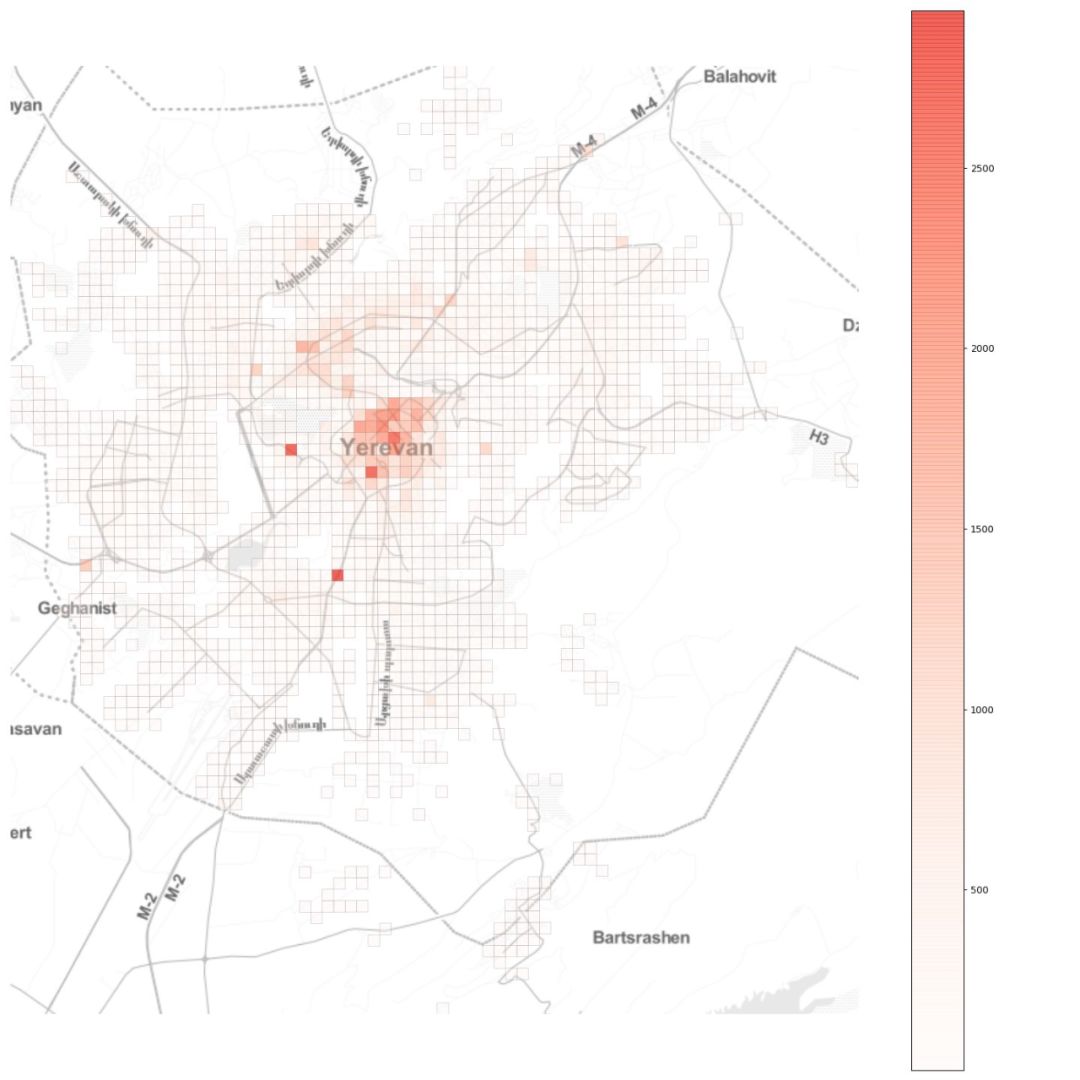
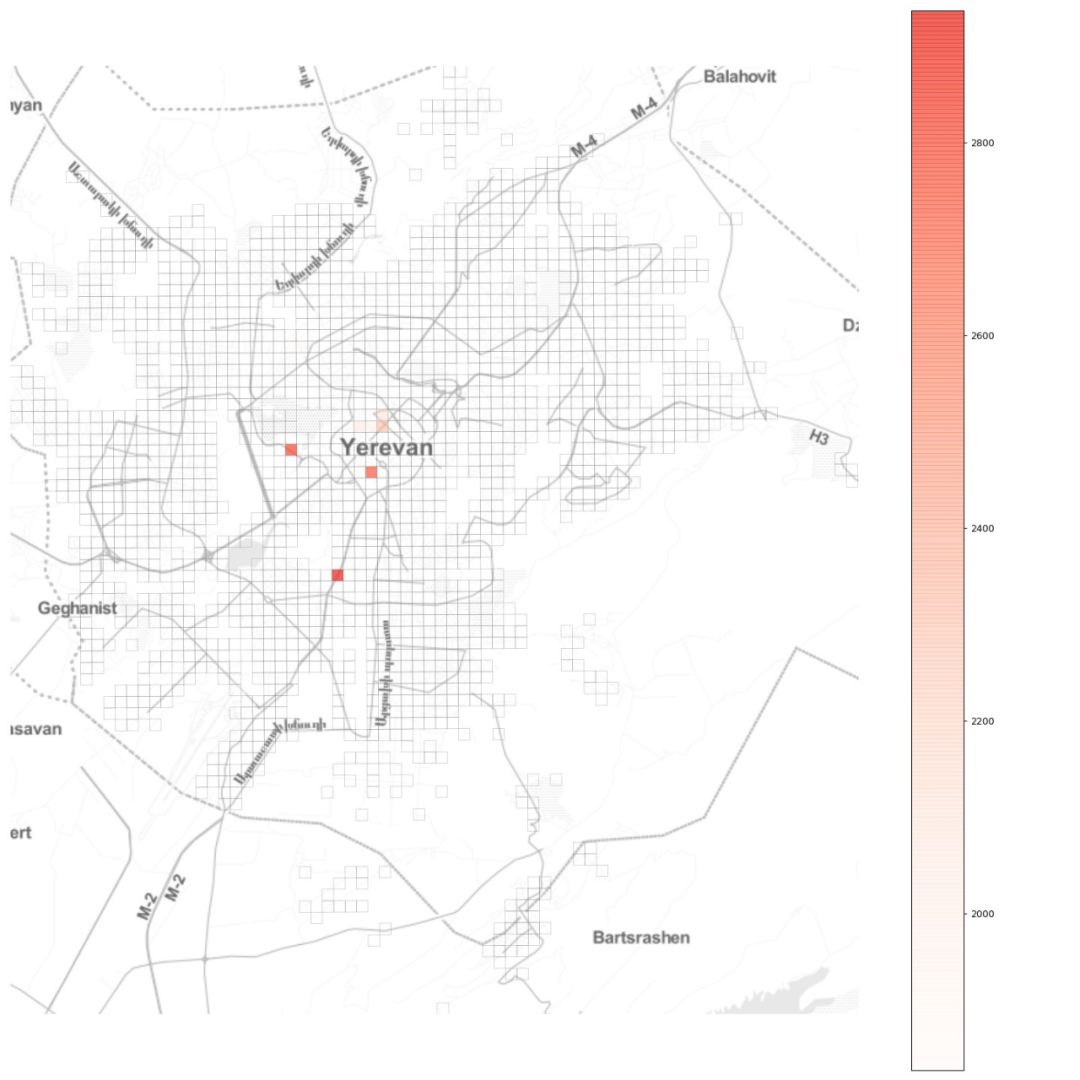
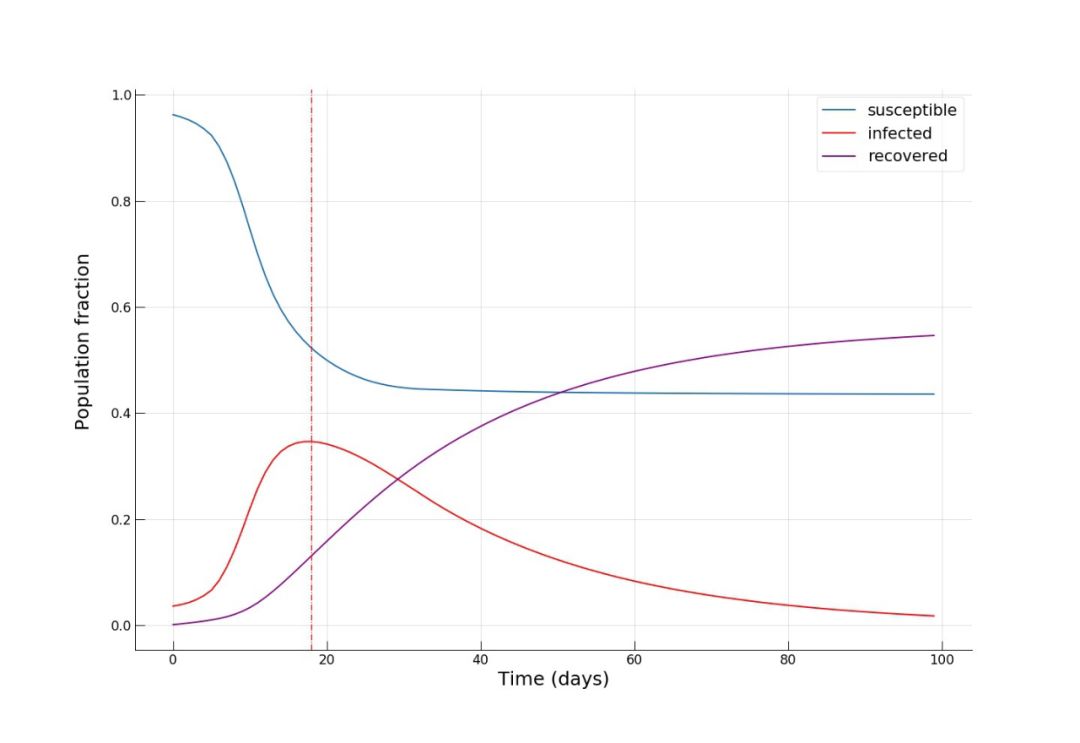
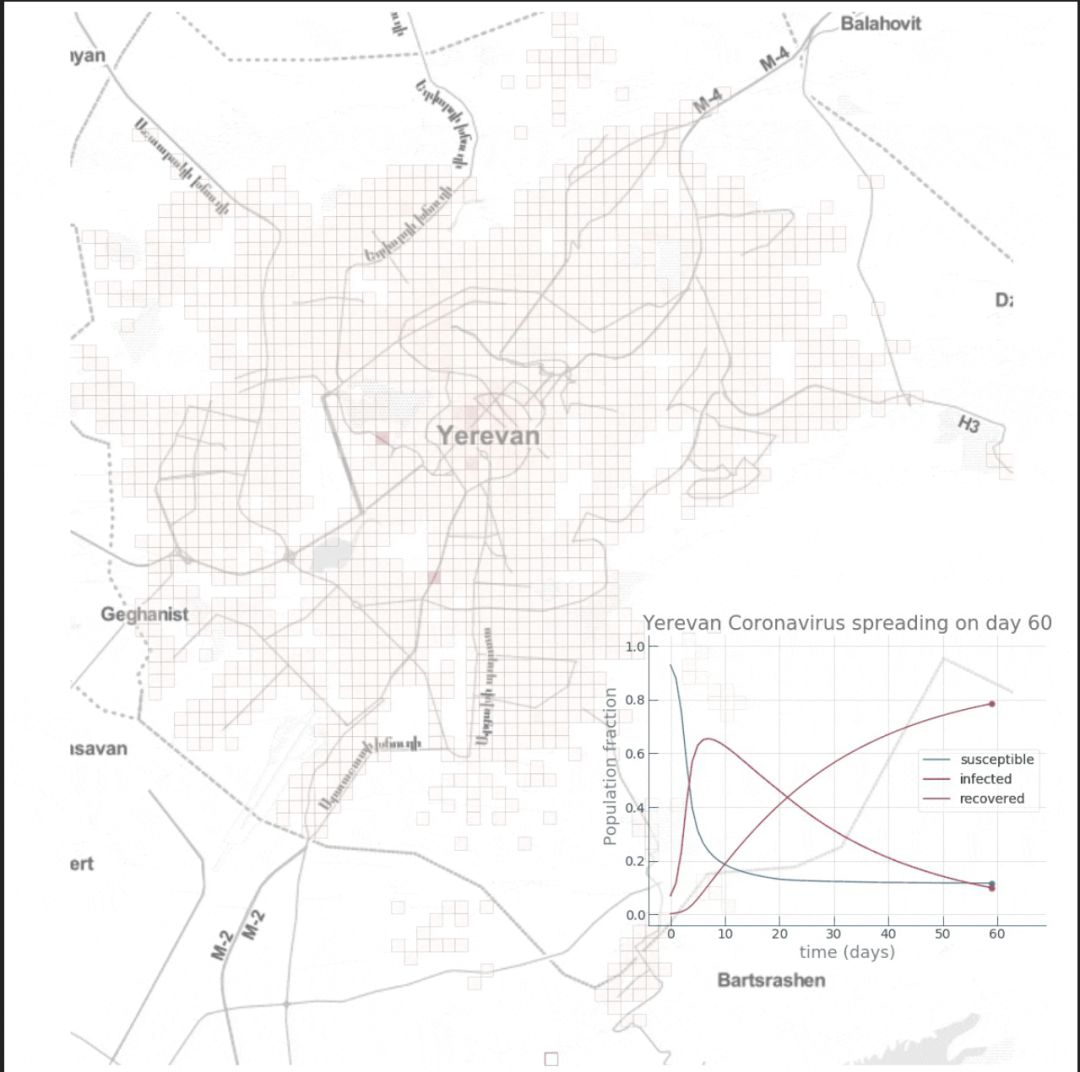
自去年12月以来,新型冠状病毒所引发的疫情已经给城市活动带来了很大影响。 怎样确切了解病毒的传播过程,从而帮助城市更好提出措施? 使用建模的方法也能起到一些作用。 本文是一篇Python教程,教你在家中也可以建模疫情传播。 本文以亚美尼亚共和国首都埃里温作为案例,对冠状病毒在该城市中的蔓延情况进行数学建模和模拟,并观察城市流动模式对病毒传播的影响。 读者也可根据文末的示例代码,自己上手使用。
S_i,t:尚未感染或易感人群(易感组);
I_i,t:感染该疾病且具备传染性的人口(感染组);
R_i,t:感染疾病后,由于痊愈或死亡从感染组移出的人口。这组人不再感染病毒,也不会将病毒传播给他人。
import numpy as np
# initialize the population vector from the origin-destination flow matrix
N_k = np.abs(np.diagonal(OD) + OD.sum(axis=0) - OD.sum(axis=1))
locs_len = len(N_k) # number of locations
SIR = np.zeros(shape=(locs_len, 3)) # make a numpy array with 3 columns for keeping track of the S, I, R groups
SIR[:,0] = N_k # initialize the S group with the respective populations
first_infections = np.where(SIR[:, 0]<=thresh, SIR[:, 0]//20, 0) # for demo purposes, randomly introduce infections
SIR[:, 0] = SIR[:, 0] - first_infections
SIR[:, 1] = SIR[:, 1] + first_infections # move infections to the I group
# row normalize the SIR matrix for keeping track of group proportions
row_sums = SIR.sum(axis=1)
SIR_n = SIR / row_sums[:, np.newaxis]
# initialize parameters
beta = 1.6
gamma = 0.04
public_trans = 0.5 # alpha
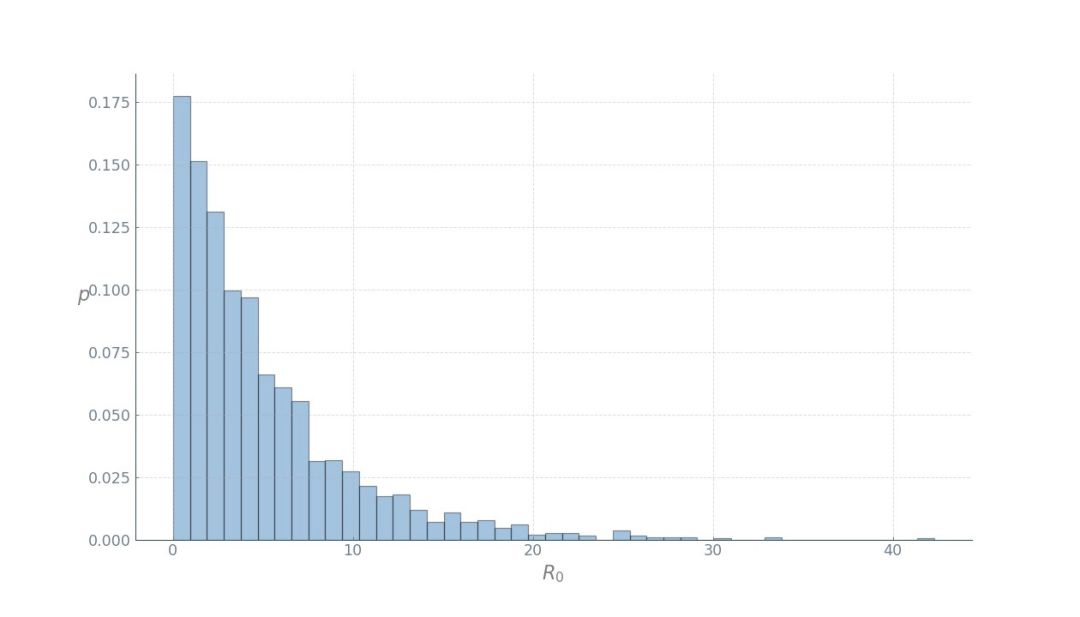
R0 = beta/gamma
beta_vec = np.random.gamma(1.6, 2, locs_len)
gamma_vec = np.full(locs_len, gamma)
public_trans_vec = np.full(locs_len, public_trans)
# make copy of the SIR matrices
SIR_sim = SIR.copy()
SIR_nsim = SIR_n.copy()
# run model
print(SIR_sim.sum(axis=0).sum() == N_k.sum())
from tqdm import tqdm_notebook
infected_pop_norm = []
susceptible_pop_norm = []
recovered_pop_norm = []
for time_step in tqdm_notebook(range(100)):
infected_mat = np.array([SIR_nsim[:,1],]*locs_len).transpose()
OD_infected = np.round(OD*infected_mat)
inflow_infected = OD_infected.sum(axis=0)
inflow_infected = np.round(inflow_infected*public_trans_vec)
print('total infected inflow: ', inflow_infected.sum())
new_infect = beta_vec*SIR_sim[:, 0]*inflow_infected/(N_k + OD.sum(axis=0))
new_recovered = gamma_vec*SIR_sim[:, 1]
new_infect = np.where(new_infect>SIR_sim[:, 0], SIR_sim[:, 0], new_infect)
SIR_sim[:, 0] = SIR_sim[:, 0] - new_infect
SIR_sim[:, 1] = SIR_sim[:, 1] + new_infect - new_recovered
SIR_sim[:, 2] = SIR_sim[:, 2] + new_recovered
SIR_sim = np.where(SIR_sim<0,0,SIR_sim)
# recompute the normalized SIR matrix
row_sums = SIR_sim.sum(axis=1)
SIR_nsim = SIR_sim / row_sums[:, np.newaxis]
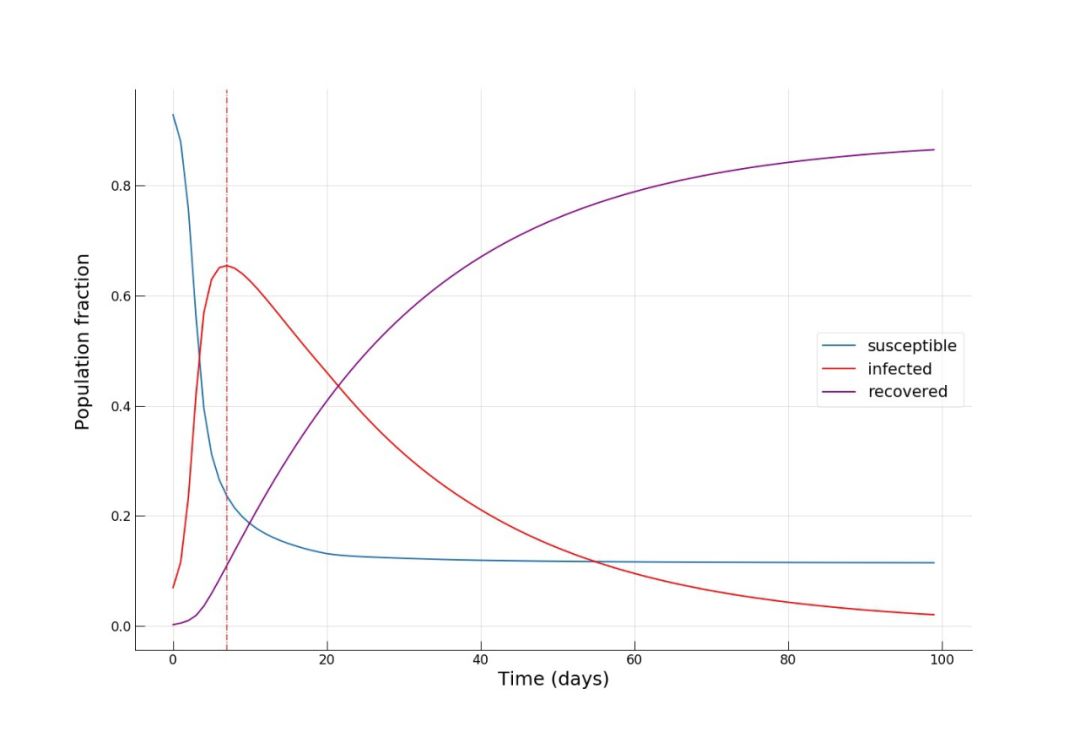
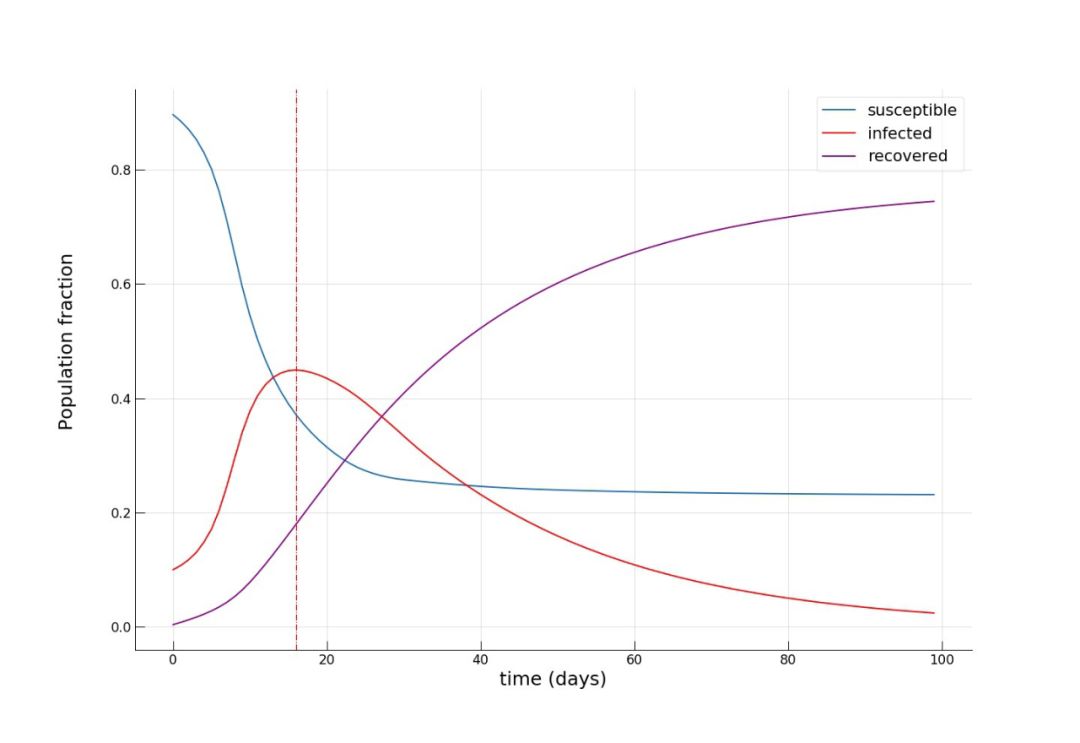
S = SIR_sim[:,0].sum()/N_k.sum()
I = SIR_sim[:,1].sum()/N_k.sum()
R = SIR_sim[:,2].sum()/N_k.sum()
print(S, I, R, (S+I+R)*N_k.sum(), N_k.sum())
print('\n')
infected_pop_norm.append(I)
susceptible_pop_norm.append(S)
recovered_pop_norm.append(R)原文地址:https://towardsdatascience.com/modelling-the-coronavirus-epidemic-spreading-in-a-city-with-python-babd14d82fa2
登录查看更多
相关内容
Arxiv
11+阅读 · 2018年5月16日
Arxiv
13+阅读 · 2018年1月25日