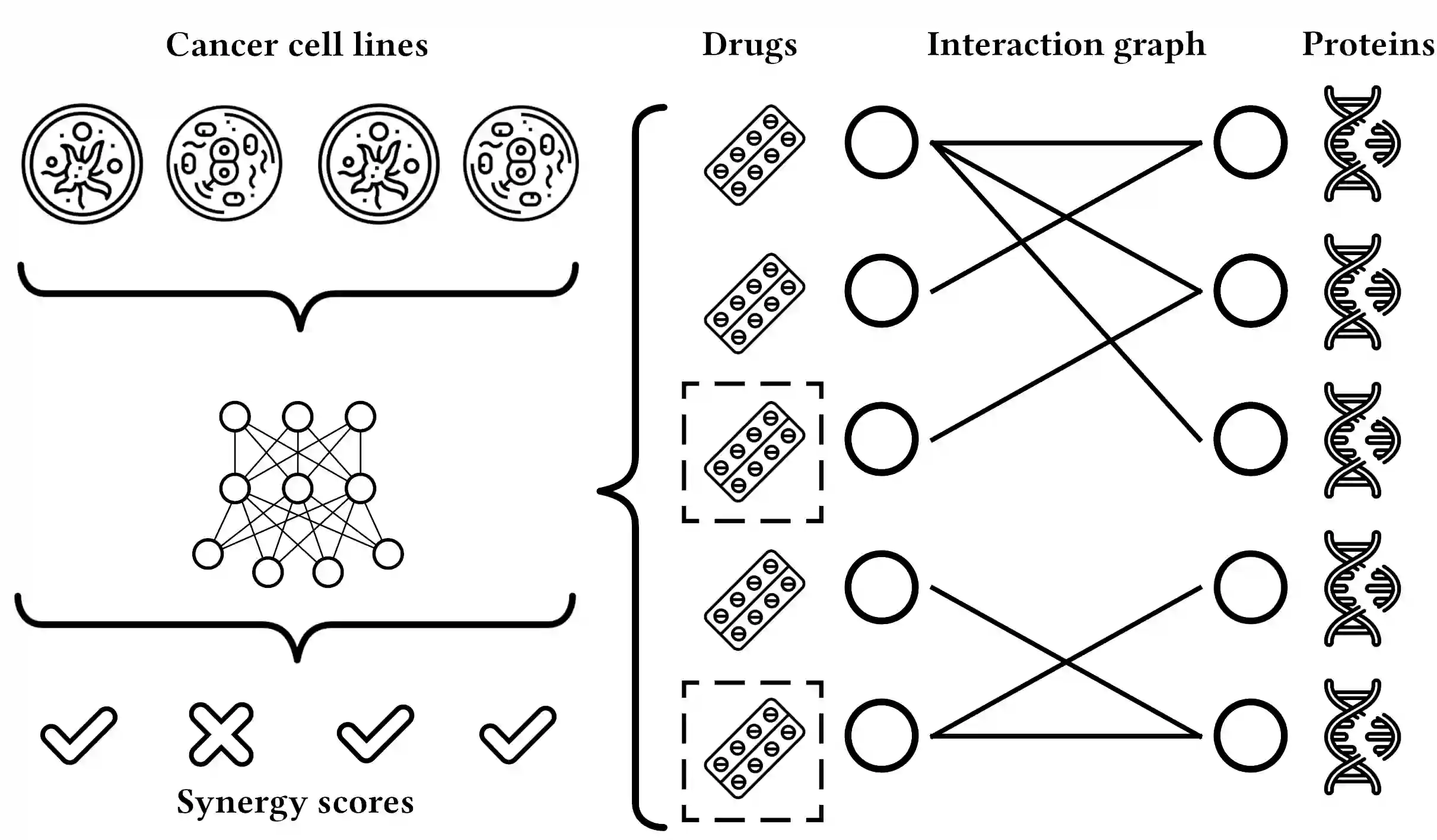
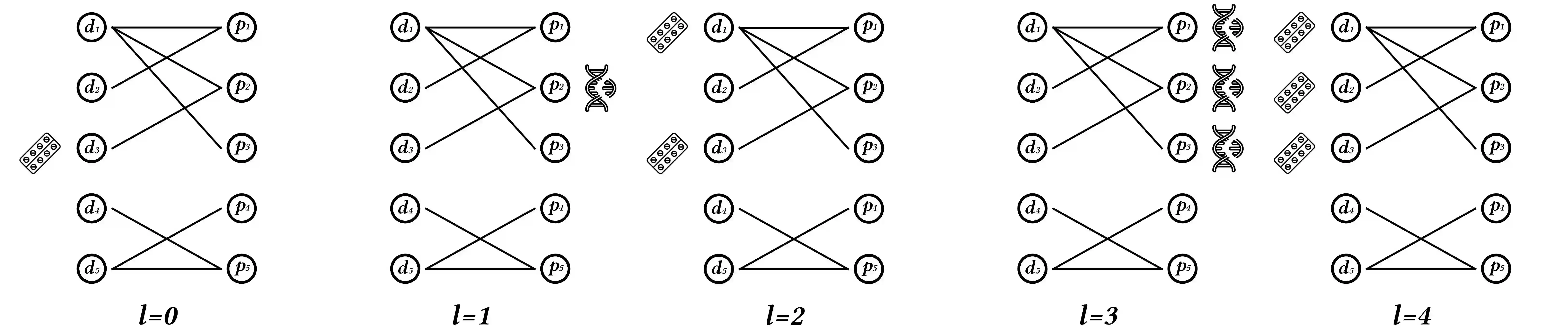
We propose the molecular omics network (MOOMIN) a multimodal graph neural network that can predict the synergistic effect of drug combinations for cancer treatment. Our model captures the representation based on the context of drugs at multiple scales based on a drug-protein interaction network and metadata. Structural properties of the compounds and proteins are encoded to create vertex features for a message-passing scheme that operates on the bipartite interaction graph. Propagated messages form multi-resolution drug representations which we utilized to create drug pair descriptors. By conditioning the drug combination representations on the cancer cell type we define a synergy scoring function that can inductively score unseen pairs of drugs. Experimental results on the synergy scoring task demonstrate that MOOMIN outperforms state-of-the-art graph fingerprinting, proximity preserving node embedding, and existing deep learning approaches. Further results establish that the predictive performance of our model is robust to hyperparameter changes. We demonstrate that the model makes high-quality predictions over a wide range of cancer cell line tissues, out-of-sample predictions can be validated with external synergy databases, and that the proposed model is data-efficient at learning.
翻译:我们建议分子粒子网络(MOOMIN)是一个多式图形神经网络,可以预测药物组合对癌症治疗的协同效应。我们的模型根据药物-蛋白互动网络和元数据,捕捉基于多种比例的药物在多种规模上的代表性。化合物和蛋白质的结构特性被编码,为双面互动图上运行的信息传递计划创建顶点特征。预发信息形成多种分辨率药物表示,我们用来创建药物配对描述器。通过调整癌症细胞类型的药物组合表现,我们定义了一种协同作用评分功能,能够感应到看不见的一对药物。协同增效评分任务的实验结果表明,MOOMIN优于最新图形指纹、近距离保存节点嵌入和现有的深层学习方法。进一步的结果证明,我们模型的预测性能强于超分度变化。我们证明模型对一系列广泛的癌症细胞细胞组织进行高质量的预测,外模模值预测可以与外部协同性数据库验证,拟议的数据可以与外部协同性学习。