哈工大硕士生用 Python 实现了 11 种经典数据降维算法,源代码库已开放
新智元推荐
【新智元导读】网上关于各种降维算法的资料参差不齐,同时大部分不提供源代码。这里有个 GitHub 项目整理了使用 Python 实现了 11 种经典的数据抽取(数据降维)算法,包括:PCA、LDA、MDS、LLE、TSNE 等,并附有相关资料、展示效果;非常适合机器学习初学者和刚刚入坑数据挖掘的小伙伴。来 新智元AI朋友圈 和AI大咖们一起讨论吧。
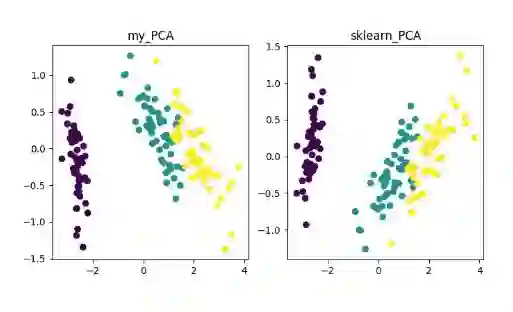
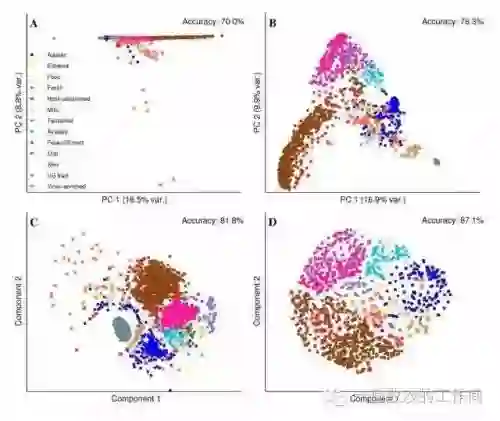
网上关于各种降维算法的资料参差不齐,同时大部分不提供源代码。这里有个 GitHub 项目整理了使用 Python 实现了 11 种经典的数据抽取(数据降维)算法,包括:PCA、LDA、MDS、LLE、TSNE 等,并附有相关资料、展示效果;非常适合机器学习初学者和刚刚入坑数据挖掘的小伙伴。
-
使得数据集更易使用 -
确保变量之间彼此独立 -
降低算法计算运算成本 -
去除噪音
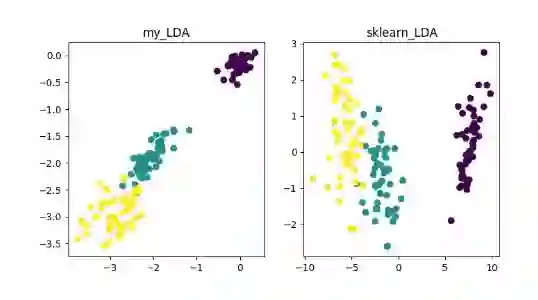
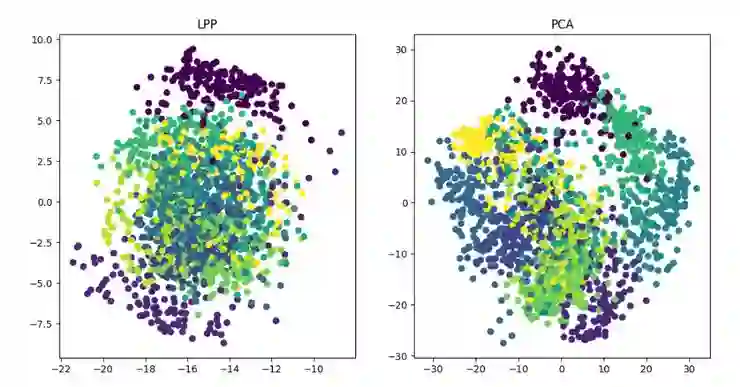
线性降维方法:PCA 、ICA LDA、LFA、LPP(LE 的线性表示)
非线性降维方法:
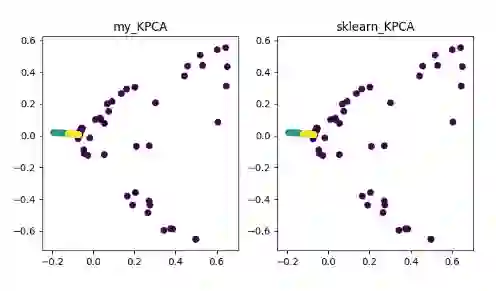
基于核函数的非线性降维方法——KPCA 、KICA、KDA
基于特征值的非线性降维方法(流型学习)——ISOMAP、LLE、LE、LPP、LTSA、MVU
-
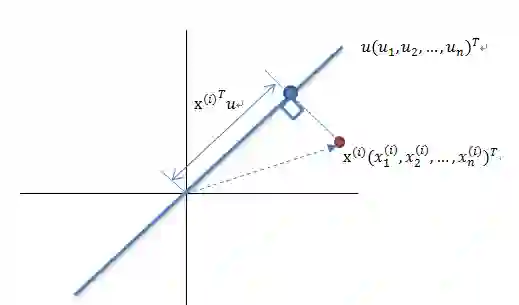
算法输入:数据集 Xmxn; -
按列计算数据集 X 的均值 Xmean,然后令 Xnew=X−Xmean; -
求解矩阵 Xnew 的协方差矩阵,并将其记为 Cov; -
计算协方差矩阵 COv 的特征值和相应的特征向量; -
将特征值按照从大到小的排序,选择其中最大的 k 个,然后将其对应的 k 个特征向量分别作为列向量组成特征向量矩阵 Wnxk; -
计算 XnewW,即将数据集 Xnew 投影到选取的特征向量上,这样就得到了我们需要的已经降维的数据集 XnewW。
详细步骤可参考《从零开始实现主成分分析 (PCA) 算法》: https://blog.csdn.net/u013719780/article/details/78352262
关于 PCA 算法的代码如下:
from __future__ import print_functionfrom sklearn import datasetsimport matplotlib.pyplot as pltimport matplotlib.cm as cmximport matplotlib.colors as colorsimport numpy as np%matplotlib inlinedef shuffle_data(X, y, seed=None):if seed:np.random.seed(seed)idx = np.arange(X.shape[0])np.random.shuffle(idx)return X[idx], y[idx]# 正规化数据集 Xdef normalize(X, axis=-1, p=2):lp_norm = np.atleast_1d(np.linalg.norm(X, p, axis))lp_norm[lp_norm == 0] = 1return X / np.expand_dims(lp_norm, axis)# 标准化数据集 Xdef standardize(X):X_std = np.zeros(X.shape)mean = X.mean(axis=0)std = X.std(axis=0)# 做除法运算时请永远记住分母不能等于 0 的情形# X_std = (X - X.mean(axis=0)) / X.std(axis=0)for col in range(np.shape(X)[1]):if std[col]:X_std[:, col] = (X_std[:, col] - mean[col]) / std[col]return X_std# 划分数据集为训练集和测试集def train_test_split(X, y, test_size=0.2, shuffle=True, seed=None):if shuffle:X, y = shuffle_data(X, y, seed)n_train_samples = int(X.shape[0] * (1-test_size))x_train, x_test = X[:n_train_samples], X[n_train_samples:]y_train, y_test = y[:n_train_samples], y[n_train_samples:]return x_train, x_test, y_train, y_test# 计算矩阵 X 的协方差矩阵def calculate_covariance_matrix(X, Y=np.empty((0,0))):if not Y.any():Y = Xn_samples = np.shape(X)[0]covariance_matrix = (1 / (n_samples-1)) * (X - X.mean(axis=0)).T.dot(Y - Y.mean(axis=0))return np.array(covariance_matrix, dtype=float)# 计算数据集 X 每列的方差def calculate_variance(X):n_samples = np.shape(X)[0]variance = (1 / n_samples) * np.diag((X - X.mean(axis=0)).T.dot(X - X.mean(axis=0)))return variance# 计算数据集 X 每列的标准差def calculate_std_dev(X):std_dev = np.sqrt(calculate_variance(X))return std_dev# 计算相关系数矩阵def calculate_correlation_matrix(X, Y=np.empty([0])):# 先计算协方差矩阵covariance_matrix = calculate_covariance_matrix(X, Y)# 计算 X, Y 的标准差std_dev_X = np.expand_dims(calculate_std_dev(X), 1)std_dev_y = np.expand_dims(calculate_std_dev(Y), 1)correlation_matrix = np.divide(covariance_matrix, std_dev_X.dot(std_dev_y.T))return np.array(correlation_matrix, dtype=float)class PCA():"""主成份分析算法 PCA,非监督学习算法."""def __init__(self):self.eigen_values = Noneself.eigen_vectors = Noneself.k = 2def transform(self, X):"""将原始数据集 X 通过 PCA 进行降维"""covariance = calculate_covariance_matrix(X)# 求解特征值和特征向量self.eigen_values, self.eigen_vectors = np.linalg.eig(covariance)# 将特征值从大到小进行排序,注意特征向量是按列排的,即 self.eigen_vectors 第 k 列是 self.eigen_values 中第 k 个特征值对应的特征向量idx = self.eigen_values.argsort()[::-1]eigenvalues = self.eigen_values[idx][:self.k]eigenvectors = self.eigen_vectors[:, idx][:, :self.k]# 将原始数据集 X 映射到低维空间X_transformed = X.dot(eigenvectors)return X_transformeddef main():# Load the datasetdata = datasets.load_iris()X = data.datay = data.target# 将数据集 X 映射到低维空间X_trans = PCA().transform(X)x1 = X_trans[:, 0]x2 = X_trans[:, 1]cmap = plt.get_cmap('viridis')colors = [cmap(i) for i in np.linspace(0, 1, len(np.unique(y)))]class_distr = []# Plot the different class distributionsfor i, l in enumerate(np.unique(y)):_x1 = x1[y == l]_x2 = x2[y == l]_y = y[y == l]class_distr.append(plt.scatter(_x1, _x2, color=colors[i]))# Add a legendplt.legend(class_distr, y, loc=1)# Axis labelsplt.xlabel('Principal Component 1')plt.ylabel('Principal Component 2')plt.show()if __name__ == "__main__":main()
KPCA(kernel PCA)
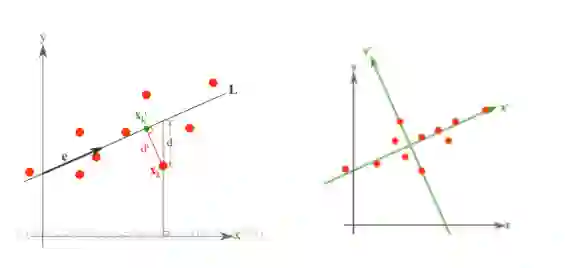
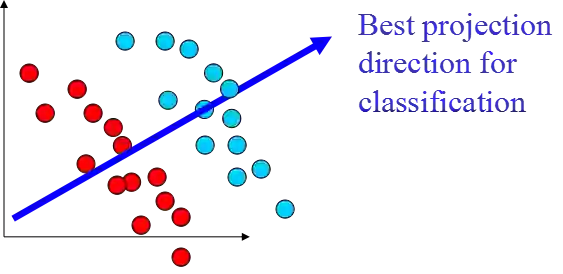
LDA(Linear Discriminant Analysis)
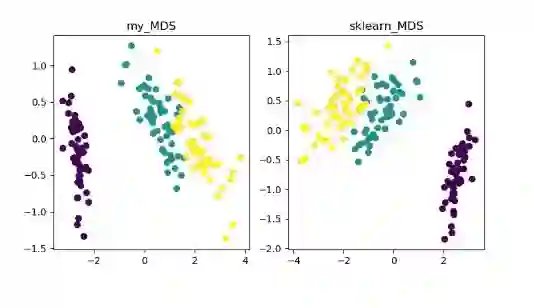
MDS(multidimensional scaling)
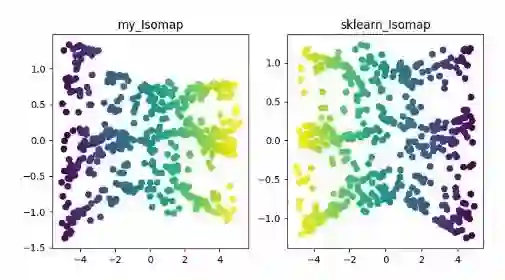
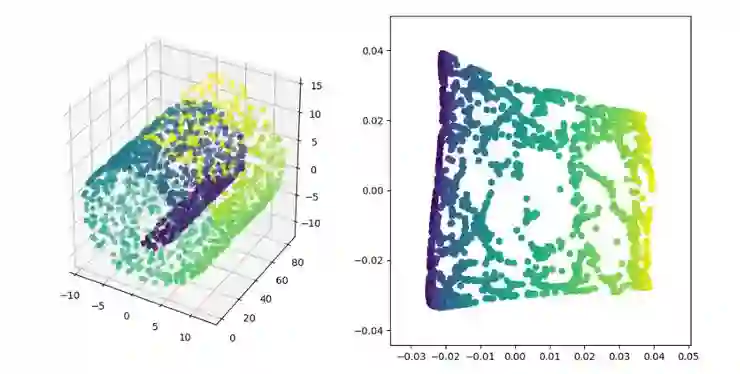
ISOMAP
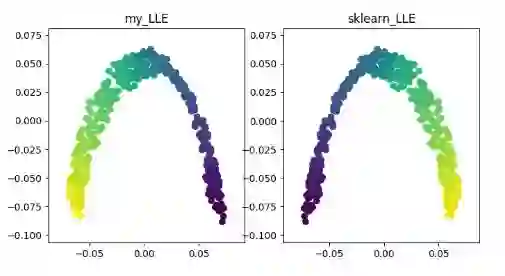
LLE(locally linear embedding)
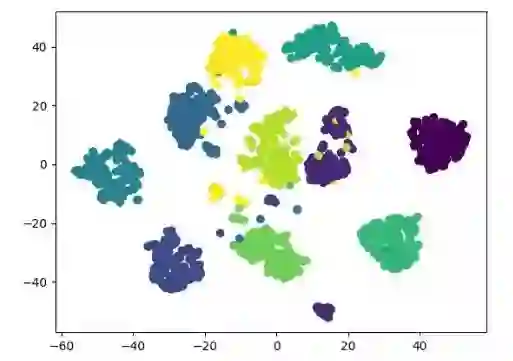
t-SNE
LE(Laplacian Eigenmaps)
LPP(Locality Preserving Projections)
Github 项目地址:

登录查看更多